 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010546341.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1075aa MW: 119660 Da PI: 5.7662 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 188.6 | 6.2e-59 | 19 | 136 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+e ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgkt++E+hekLKvg+v+vl+cyYah+e+n++fq
XP_010546341.1 19 LSEaQHRWLRPAEICEILRNYQKFHIASEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTIKEAHEKLKVGSVDVLHCYYAHGEDNENFQ 113
45559****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcyw+Le+++ +iv+vhylevk
XP_010546341.1 114 RRCYWMLEQDFMHIVFVHYLEVK 136
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 86.507 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-86 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.7E-52 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 4.9E-5 | 454 | 557 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 2.52E-15 | 481 | 567 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 3.0E-19 | 674 | 779 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.23E-18 | 674 | 775 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.27E-16 | 675 | 773 | No hit | No description |
| Pfam | PF12796 | 1.9E-7 | 675 | 741 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.571 | 681 | 775 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 1600 | 681 | 710 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.244 | 681 | 713 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.327 | 714 | 746 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0028 | 714 | 743 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.55 | 889 | 911 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 9.82E-8 | 889 | 940 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.84 | 891 | 919 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 9.7E-4 | 891 | 910 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 912 | 934 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.395 | 913 | 937 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.7E-4 | 915 | 934 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1075 aa Download sequence Send to blast |
MADRGSFGFG PRLDIGQLLS EAQHRWLRPA EICEILRNYQ KFHIASEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTIKEAHEK LKVGSVDVLH CYYAHGEDNE NFQRRCYWML 120 EQDFMHIVFV HYLEVKGNRT SSGGMKENNS NSINGTTSVN IDSTASPTST LSSLCEDADS 180 GDSHQASSSL QAFQDPKITT TQIRDTPDAG TSNSYYMGSF SGTREGWTSA AGMGTVSPFY 240 GNSVRESDSQ RSGDVSAWGA AFEDSTARHH NLPFNDLVTQ LPPSTTELLH VHGVENAGKG 300 ELFAADNGSK ELLRNPLQNQ VNWQIPVQDN LPLPNWPIDL VSQTGMGDDT DLTVFEQGAH 360 DNFETFSSLL NIRSQQSLGN GVHFPLPSVE SEYTPKLNPD ALQYETSTSL MLPRKPFLSK 420 EDSLKKVDSF SRWASKELGE MEDLQMQPSS SGGIAWTTVE CETAAAGSCL SPSLSQDQRF 480 TIIDFWPKWG HTDSEVEVMV IGTFLMNKQE VTKYNWSCMF GEVEVPAEIL VDGVLCCHAP 540 PHTVGQVSFY ITCSDRFACS EVREFDFLPG STKNLNVSDV YGTSTIEASL YMRFERLLAY 600 RSPVIERHIF EDVGDKRKHI SKIMLLKDER ESFFPGTNQK DSIEQEAKGH VIRDLFEEKL 660 YTWLIHKVTE DVKGPNILDE GGQGVLHFAA ALGYDWVIKP ILAAGVSINF RDANGWTALH 720 WAAFSGREQT VAVLVSLGAD AGALTDPSPE FPLGKTAADL ASGNGHKGIS GFLAESSLTS 780 YLEKLAMNES KENSSANSSG AKVVQTISER TATPMSYGDV PEALSLKDSL TAVCNATQAA 840 DRIHQVFRMQ SFQRKQLSQF GNEESDISDE LAVSFATAKT GKPGLNDGVV HAAAVNIQKK 900 YRGWKKRKEF LLIRQRIVKI QAHVRGHQVR KQYRTVIWSV GILEKVILRW RRKGSGLRGF 960 RRDAIAKQAK PETPVPPQEK EDDDYDFLKE GRKQTEERLQ KALTRVKSMV QYPEARAQYR 1020 RLLTVVEGFR ESEASSSGGA NTAEEEAAAA ANTDDDLLDI DSLLDDDTFM SIAFE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
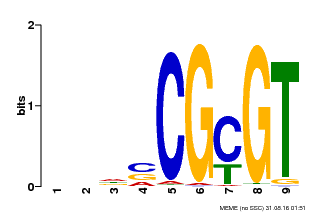
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010546341.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010546341.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2-like isoform X1 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | D7MR88 | 0.0 | D7MR88_ARALL; Calmodulin-binding protein | ||||
| STRING | XP_010546341.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



