 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010547306.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 264aa MW: 29905.4 Da PI: 8.9919 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 90.6 | 8.2e-29 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien nrqvtfskRr+g+lKKA EL vLCdaevavi+fs++gkl+e+ss
XP_010547306.1 9 KRIENANNRQVTFSKRRAGLLKKARELAVLCDAEVAVIVFSNSGKLFEFSS 59
79***********************************************96 PP
| |||||||
| 2 | K-box | 74.4 | 3.2e-25 | 73 | 172 | 2 | 100 |
K-box 2 qkssgks.leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLr 95
qkssg e + + + e++ Lk+ei++L+++q +llG++L+ LslkeLq Le+qL++ l ++R++K++llleq+ee++ ke+++ en++Lr
XP_010547306.1 73 QKSSGPPpIECKAEVDNCIEVELLKEEISKLREKQSQLLGKGLNGLSLKELQLLEEQLNETLLSVRERKEQLLLEQLEESRLKEQRAMLENETLR 167
6666666644555667789**************************************************************************** PP
K-box 96 kklee 100
+++ee
XP_010547306.1 168 RQVEE 172
**987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 9.1E-43 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 32.941 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 6.54E-33 | 2 | 74 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 4.78E-44 | 2 | 76 | No hit | No description |
| PRINTS | PR00404 | 7.6E-30 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 2.9E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.6E-30 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.6E-30 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 13.771 | 86 | 176 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 7.6E-17 | 90 | 170 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010262 | Biological Process | somatic embryogenesis | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048577 | Biological Process | negative regulation of short-day photoperiodism, flowering | ||||
| GO:0060862 | Biological Process | negative regulation of floral organ abscission | ||||
| GO:0060867 | Biological Process | fruit abscission | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 264 aa Download sequence Send to blast |
MGRGKIEIKR IENANNRQVT FSKRRAGLLK KARELAVLCD AEVAVIVFSN SGKLFEFSSS 60 GMKKTLSRYN KYQKSSGPPP IECKAEVDNC IEVELLKEEI SKLREKQSQL LGKGLNGLSL 120 KELQLLEEQL NETLLSVRER KEQLLLEQLE ESRLKEQRAM LENETLRRQV EELRGLLPSA 180 NLRYFPSYIE CYPVEIKKPP VKDADFGGFN CMAQKTDSDT TLQLGLPGEA EERRKRKEPE 240 KGSSSGDSVL TNTARACIQR ISLV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6byy_A | 1e-23 | 1 | 88 | 1 | 83 | MEF2 CHIMERA |
| 6byy_B | 1e-23 | 1 | 88 | 1 | 83 | MEF2 CHIMERA |
| 6byy_C | 1e-23 | 1 | 88 | 1 | 83 | MEF2 CHIMERA |
| 6byy_D | 1e-23 | 1 | 88 | 1 | 83 | MEF2 CHIMERA |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 232 | 237 | RRKRKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00508 | DAP | Transfer from AT5G13790 | Download |
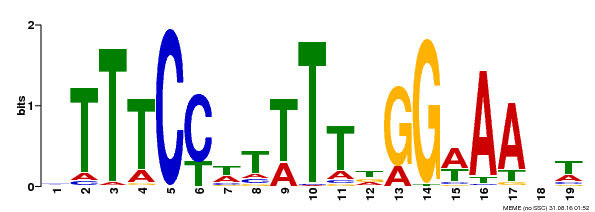
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010547306.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010547306.1 | 0.0 | PREDICTED: agamous-like MADS-box protein AGL15 isoform X4 | ||||
| Swissprot | Q39295 | 1e-109 | AGL15_BRANA; Agamous-like MADS-box protein AGL15 | ||||
| TrEMBL | R0FJ29 | 1e-112 | R0FJ29_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010547302.1 | 1e-175 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13790.1 | 1e-64 | AGAMOUS-like 15 | ||||



