 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010549420.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 249aa MW: 28133.8 Da PI: 5.2842 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 96.8 | 1.4e-30 | 71 | 129 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
+D ++WrKYGqK++kgs++pr+YYrC+s+ gCp++k+ver+++dp+++ +tY+ eHnh+
XP_010549420.1 71 SDSWAWRKYGQKPIKGSPYPRGYYRCSSSkGCPARKQVERNRDDPTMILVTYTCEHNHP 129
69*************************998****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.7E-32 | 62 | 131 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.592 | 65 | 131 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 8.89E-27 | 66 | 131 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.7E-35 | 70 | 130 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.0E-26 | 72 | 129 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 249 aa Download sequence Send to blast |
MKRALEMERS SFSESSQETG PGSPTSSIFN DMKLISSPSP KRRRSVQKRL VTVPIKDVEG 60 SRMKGETAPP SDSWAWRKYG QKPIKGSPYP RGYYRCSSSK GCPARKQVER NRDDPTMILV 120 TYTCEHNHPW PASKTGPIKH RPKPEPEPEP EIVPELELEP EQSKFMDLGE IGTTPSCVDE 180 FAWFGEMETT SSTILESPIF SSDKKTVPPP ENADVAMFFP MREEDESLFA DLGELPECSA 240 VFRRPLEIY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5w3x_B | 9e-20 | 48 | 131 | 6 | 79 | Disease resistance protein RRS1 |
| 5w3x_D | 9e-20 | 48 | 131 | 6 | 79 | Disease resistance protein RRS1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00167 | DAP | Transfer from AT1G29280 | Download |
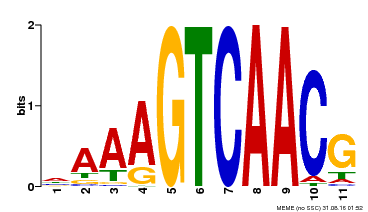
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010549420.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010549420.1 | 0.0 | PREDICTED: probable WRKY transcription factor 65 isoform X2 | ||||
| Swissprot | Q9LP56 | 1e-111 | WRK65_ARATH; Probable WRKY transcription factor 65 | ||||
| TrEMBL | A0A178W6S1 | 1e-110 | A0A178W6S1_ARATH; WRKY65 | ||||
| STRING | XP_010549419.1 | 1e-166 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G29280.1 | 1e-114 | WRKY DNA-binding protein 65 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



