 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010549755.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | NF-YB | ||||||||
| Protein Properties | Length: 134aa MW: 15255.8 Da PI: 4.8783 | ||||||||
| Description | NF-YB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NF-YB | 164.2 | 1.8e-51 | 2 | 97 | 2 | 97 |
NF-YB 2 reqdrflPianvsrimkkvlPanakiskdaketvqecvsefisfvtseasdkcqrekrktingddllwalatlGfedyveplkvylkkyrelegek 97
+++dr+lPianv+rimk++lP nakisk+ak+tvqec++efisfvt+easdkc++e+rkt+ngdd++wal+tlG+++y++++ yl++yre e+e+
XP_010549755.1 2 TDEDRLLPIANVGRIMKQILPPNAKISKEAKQTVQECATEFISFVTCEASDKCHKENRKTVNGDDICWALSTLGLDNYADAMGRYLHNYRESERER 97
689******************************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.20.10 | 1.6E-49 | 2 | 120 | IPR009072 | Histone-fold |
| SuperFamily | SSF47113 | 4.19E-38 | 4 | 120 | IPR009072 | Histone-fold |
| Pfam | PF00808 | 1.7E-26 | 7 | 71 | IPR003958 | Transcription factor CBF/NF-Y/archaeal histone domain |
| PRINTS | PR00615 | 2.1E-15 | 35 | 53 | No hit | No description |
| PROSITE pattern | PS00685 | 0 | 38 | 54 | IPR003956 | Transcription factor, NFYB/HAP3, conserved site |
| PRINTS | PR00615 | 2.1E-15 | 54 | 72 | No hit | No description |
| PRINTS | PR00615 | 2.1E-15 | 73 | 91 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 134 aa Download sequence Send to blast |
MTDEDRLLPI ANVGRIMKQI LPPNAKISKE AKQTVQECAT EFISFVTCEA SDKCHKENRK 60 TVNGDDICWA LSTLGLDNYA DAMGRYLHNY RESERERAEQ NRTDDEEPNS RSELQNQQIE 120 GGRVFEKGSS SSRN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4g91_B | 9e-40 | 1 | 92 | 1 | 92 | Transcription factor HapC (Eurofung) |
| 4g92_B | 9e-40 | 1 | 92 | 1 | 92 | Transcription factor HapC (Eurofung) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the NF-Y/HAP transcription factor complex. The NF-Y complex stimulates the transcription of various genes by recognizing and binding to a CCAAT motif in promoters. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00133 | DAP | Transfer from AT1G09030 | Download |
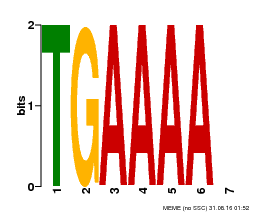
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010549755.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB493443 | 1e-83 | AB493443.1 Arabidopsis thaliana At1g09030 gene for hypothetical protein, partial cds, clone: RAAt1g09030. | |||
| GenBank | BT029363 | 1e-83 | BT029363.1 Arabidopsis thaliana At1g09030 mRNA, complete cds. | |||
| GenBank | CP002684 | 1e-83 | CP002684.1 Arabidopsis thaliana chromosome 1 sequence. | |||
| GenBank | F7G19 | 1e-83 | AC000106.1 Sequence of BAC F7G19 from Arabidopsis thaliana chromosome 1, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010549755.1 | 2e-97 | PREDICTED: nuclear transcription factor Y subunit B-4 | ||||
| Swissprot | O04027 | 7e-74 | NFYB4_ARATH; Nuclear transcription factor Y subunit B-4 | ||||
| TrEMBL | R0IG98 | 5e-74 | R0IG98_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010549755.1 | 8e-97 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM255 | 28 | 229 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09030.1 | 3e-76 | nuclear factor Y, subunit B4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



