 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010550439.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 347aa MW: 36719.5 Da PI: 8.0673 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 220.5 | 3.5e-68 | 128 | 278 | 3 | 153 |
DUF702 3 sgtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa...aeaaskrkrelkskkqsalsstklssa 94
sg++sCqdCGnqakkdCah RCR CCksrgf+C+thvkstWvpaakrrerqq+laa +++ ++++ e+ +kr+re + ++s+l +t+l+ +
XP_010550439.1 128 SGGVSCQDCGNQAKKDCAHLRCRACCKSRGFECQTHVKSTWVPAAKRRERQQHLAAIQHQPQTQTqtrGESVPKRQRE--NLASSSLVCTRLPAN 220
6889***************************************************99988777664334455555555..478899999**9998 PP
DUF702 95 eskkeletsslPeevsseavfrcvrvssvdd.geeelaYqtavsigGhvfkGiLydqGle 153
+s le+ ++P+e+ss+avfrcvrvssvd ge+++aYqtavsigGhvfkGiLydqG++
XP_010550439.1 221 TSG--LEMGQFPPELSSPAVFRCVRVSSVDVeGEDQFAYQTAVSIGGHVFKGILYDQGPD 278
765..88899********************559*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 2.8E-64 | 130 | 278 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 4.0E-26 | 132 | 174 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 2.6E-25 | 228 | 277 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 347 aa Download sequence Send to blast |
MAGFFSLGGG GGGGGGGSNQ DEHNQGTNTN PPPVTDAWRW YRNPNSPTAN AADITSSYKG 60 TLELWQHQNQ QEIIFHQQQH QQRLDLYSSA AGLGVGPSSR TQFDISGETS AAAVANRAAL 120 MMLRTGVSGG VSCQDCGNQA KKDCAHLRCR ACCKSRGFEC QTHVKSTWVP AAKRRERQQH 180 LAAIQHQPQT QTQTRGESVP KRQRENLASS SLVCTRLPAN TSGLEMGQFP PELSSPAVFR 240 CVRVSSVDVE GEDQFAYQTA VSIGGHVFKG ILYDQGPDCS GGAAESSSCG GSQPLNLITA 300 VSSSSPNIGN KGGGGSSSSG YIDPSSLHPT PINTFMAGTQ FFPNPRS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA on 5'-ACTCTAC-3' and promotes auxin homeostasis-regulating gene expression (e.g. YUC genes), as well as genes affecting stamen development, cell expansion and timing of flowering. Synergistically with other SHI-related proteins, regulates gynoecium, stamen and leaf development in a dose-dependent manner, controlling apical-basal patterning. Promotes style and stigma formation, and influences vascular development during gynoecium development. May also have a role in the formation and/or maintenance of the shoot apical meristem (SAM). {ECO:0000269|PubMed:12361963, ECO:0000269|PubMed:16740145, ECO:0000269|PubMed:16740146, ECO:0000269|PubMed:18811619, ECO:0000269|PubMed:20154152, ECO:0000269|PubMed:22318676}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
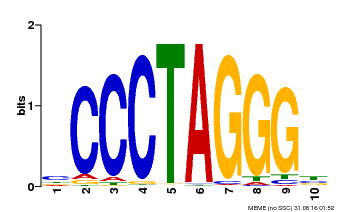
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010550439.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Regulated by ESR1 and ESR2. {ECO:0000269|PubMed:21976484}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010550439.1 | 0.0 | PREDICTED: protein SHI RELATED SEQUENCE 1-like | ||||
| Swissprot | Q9SD40 | 1e-137 | SRS1_ARATH; Protein SHI RELATED SEQUENCE 1 | ||||
| TrEMBL | A0A0D3A8F7 | 1e-145 | A0A0D3A8F7_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6EZB1 | 1e-145 | A0A3P6EZB1_BRAOL; Uncharacterized protein | ||||
| STRING | XP_010550439.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3419 | 28 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G51060.1 | 1e-113 | Lateral root primordium (LRP) protein-related | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



