 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010550712.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1489aa MW: 168570 Da PI: 8.119 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 14.6 | 9.3e-05 | 1373 | 1398 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C+ C++sFs+k +L H r+ +
XP_010550712.1 1373 YRCNmeGCTMSFSTKQQLALHKRNiC 1398
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 12.5 | 0.00044 | 1398 | 1420 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
XP_010550712.1 1398 CPvkGCGKKFFSHKYLVQHKRVH 1420
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 11.8 | 0.00073 | 1456 | 1482 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
XP_010550712.1 1456 YVCAepGCGQTFRFVSDFSRHKRKtgH 1482
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 6.0E-15 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.883 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 2.2E-13 | 21 | 54 | IPR003349 | JmjN domain |
| SuperFamily | SSF51197 | 6.04E-28 | 115 | 176 | No hit | No description |
| PROSITE profile | PS51184 | 33.752 | 196 | 365 | IPR003347 | JmjC domain |
| SMART | SM00558 | 4.5E-52 | 196 | 365 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 6.04E-28 | 214 | 384 | No hit | No description |
| Pfam | PF02373 | 1.2E-37 | 229 | 348 | IPR003347 | JmjC domain |
| SMART | SM00355 | 7.8 | 1373 | 1395 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 5.15E-6 | 1396 | 1432 | No hit | No description |
| PROSITE profile | PS50157 | 12.321 | 1396 | 1425 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0046 | 1396 | 1420 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.8E-5 | 1397 | 1424 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1398 | 1420 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.6E-9 | 1425 | 1450 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.614 | 1426 | 1455 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0014 | 1426 | 1450 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1428 | 1450 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.53E-10 | 1436 | 1481 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-10 | 1451 | 1479 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.62 | 1456 | 1482 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.032 | 1456 | 1487 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1458 | 1482 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1489 aa Download sequence Send to blast |
MAVSEASQDV FPWLKSLPLA PEFRPSPAEF QDPIAYIFKI EKEASKYGIC KILPPVPPAS 60 KKTMISNLNR SLAARAAAAA REGGACDYDR PTFTTRQQQI GFCARKQRPA QRPVWQSGEH 120 YTFQEFEAKA KNFEKNYLKK CGKKGPLSAL EIETLYWRAT VDKPFSVEYA NDMPGSAFIP 180 LSTAAVRRRE SGDGGTVGET TWNMRAVSRA EGSLLKFMKE EIPGVTSPMV YIAMMFSWFA 240 WHVEDHDLHS LNYLHMGAGK TWYGVPKDAA VAFEEVVRVH GYGGELNPLV TFATLGEKTT 300 VMSPEVFVKA GIPCCRLVQN PGEFVVTFPR AYHSGFSHGF NCGEASNIAT PEWLTMAKDA 360 AIRRAAINYP PMVSHFQLLY DFALALCSRV PASLTTKPRS SRLKDKKKSE GERLTKELFV 420 QNVIKNNKLL HSLGRGSLVV LLPRSSSDIS VCSDLRVGSQ LRTNQEKTLQ VFRDDVKPEE 480 LGSSYGMIGF NNDTKLDKGT VSVKEKFTSL CERNRNFPSW GNNTQGTLID ARRERGDRAE 540 GLSDQRLFSC VTCGILSFDC VAIVQPREAA ARYLMSADCS FFNDWTVGSG LASSGQDTSS 600 PVMKPVAQGT GKRDVNYLYD VPVQTGDCQM KTGNQRNGTA SKTNEEKDSG ALGLLASTYG 660 DSDSEEDNDD PDVPLSEGET NITNCSPPKK YLQDCVSQKM DCNEEAGLRQ SDLNFRTDQT 720 CDGTDEFRAQ RLACRRSNGV EVHATSSCST VSCTTEQKRL GLGEGTTSLL DMDLPFVPRS 780 NEDSSRLHVF CLEHAAEVEQ QLRPIGGVRI MLICHPEYPT IEAEAKLVAE ELEIDHQWND 840 VEFRSVTRED EGRIQSALDN EEAKAGNSDW AVKLGINLFY SAVLSRSPLY SKQMPYNSVI 900 YSAFGRSSPA SSPSKPEFPG RRSSRQKKFV VGKWCGKVWM SHQVHPFLVQ QDSEDEELDR 960 NFHSRAVMDE GVTERKPFSI LRNVTTMVAR KYCRKRKMRA KPMSRKKLTS FRTECGVSDD 1020 TSEDHPYKQQ WRASGNEEDS YFETGNTVSG DLSDQMSDQR LGRRGTKFQD EEDERSDDMS 1080 EDKYPNLKRK GILRHKGVHE FESEDEVSDR SLAEEYTARD FAPSDNSMEN GFQPNKQSRL 1140 LEMDREASDD DSADDDDDNI YRQQREIPRS KRTRIFKNVF SYDSEENGSY RHRSRMPRRT 1200 RKASRIGRKE EFSYESAEDN TDDENRRVMK HRKVKNIEEK EDERCSDSVE EQDFCSRRKR 1260 TATRKAKPEI LQSLKGTKGQ PASRKKKKKQ EGIRVLNVKQ EKDNVPLDSY TEGPSTRLRV 1320 RNQKPSRVSE TKSKKPGKKE RNASSFSRVA NEEDDDDAEE EEEDDEEEGS SVYRCNMEGC 1380 TMSFSTKQQL ALHKRNICPV KGCGKKFFSH KYLVQHKRVH LDDRPLKCPW KGCKMTFKWA 1440 WARTEHIRVH TGERPYVCAE PGCGQTFRFV SDFSRHKRKT GHSVKKTKR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 2e-76 | 9 | 453 | 4 | 413 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-76 | 9 | 453 | 4 | 413 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 994 | 1006 | KRKMRAKPMSRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
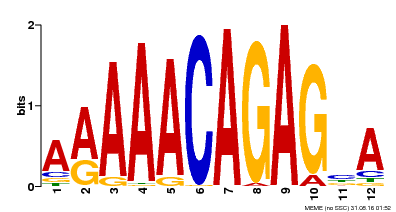
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010550712.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010550712.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A1J3JIY4 | 0.0 | A0A1J3JIY4_NOCCA; Lysine-specific demethylase REF6 | ||||
| STRING | XP_010550712.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



