 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010553611.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 558aa MW: 60998.6 Da PI: 7.1339 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 89.2 | 3.7e-28 | 214 | 267 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv+av++L G++kA+Pk+ilelm+v+gLt+e+v+SHLQk+Rl
XP_010553611.1 214 KPRVVWSVELHQQFVSAVNHL-GIDKAVPKRILELMNVPGLTRENVASHLQKFRL 267
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 76.2 | 1.2e-25 | 29 | 137 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaG 93
vl+vdD+ + +++l+++l++ y +v+ ++++ al++l+e++ +Dl+l D+ mpgmdG++ll++ e +lp+i+++a g +++ ++ G
XP_010553611.1 29 VLVVDDDASCLKILEKMLRRLMY-QVTICSQANVALAILREMKgcFDLVLSDVHMPGMDGYKLLEQVGLEM-DLPVIMMSADGRTSTVMTGINHG 121
89*********************.*******************************************6644.8********************** PP
ESEEEESS--HHHHHH CS
Response_reg 94 akdflsKpfdpeelvk 109
a d+l Kp+ e+l +
XP_010553611.1 122 ACDYLIKPIRLEALKN 137
************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 8.2E-150 | 8 | 550 | IPR017053 | Response regulator B-type, plant |
| Gene3D | G3DSA:3.40.50.2300 | 2.2E-41 | 26 | 169 | No hit | No description |
| SuperFamily | SSF52172 | 1.57E-35 | 26 | 156 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 2.6E-30 | 27 | 139 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.443 | 28 | 143 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.5E-22 | 29 | 137 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 4.58E-28 | 30 | 142 | No hit | No description |
| PROSITE profile | PS51294 | 10.954 | 211 | 270 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-29 | 211 | 272 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.15E-19 | 212 | 271 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-24 | 214 | 267 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.8E-6 | 216 | 265 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 558 aa Download sequence Send to blast |
MSGFHNGDGC SGGAAATAVS DQFPAGLRVL VVDDDASCLK ILEKMLRRLM YQVTICSQAN 60 VALAILREMK GCFDLVLSDV HMPGMDGYKL LEQVGLEMDL PVIMMSADGR TSTVMTGINH 120 GACDYLIKPI RLEALKNIWQ HVVRKKWSMN KEVHLSGALD DRDPFRQRND SPENAFSASD 180 GSDGSSKHQR KRSCVRENEE DDIEKDDLDS ASKKPRVVWS VELHQQFVSA VNHLGIDKAV 240 PKRILELMNV PGLTRENVAS HLQKFRLYLK RLSGVAPQSS VCNIAQPNLK LGRCDIQALA 300 ASGQIHPQAL AALFGQTLPV GNLHQPALVQ PTLPGPKCLP LGQSVTYGHP VAKCPPPDVS 360 KNFQHSVLLA KDHRAGFGAW LPNNHLSTMA SEKLNFQQNE DFPEDMSQKS NPTINVQPSC 420 LVVPSPSSNN CLGVTNSPAS VNQTIVGFTN AAVFDHSVLS AQQGPLSPCV SSSACTVNTD 480 NNVMPRKRHP GFVHRLPSRF AIDEMEWSLD NLDNGKNFDS EIIIKQELGV DPMNIVRSGG 540 ALILQQFHSN ELTSVFTK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 6e-24 | 210 | 273 | 1 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins (By similarity). {ECO:0000250}. | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00098 | PBM | Transfer from AT2G01760 | Download |
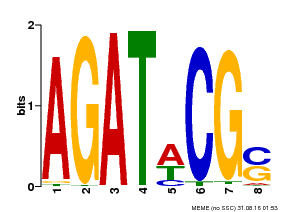
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010553611.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010553611.1 | 0.0 | PREDICTED: two-component response regulator ARR14 isoform X1 | ||||
| Swissprot | A2XE31 | 1e-121 | ORR21_ORYSI; Two-component response regulator ORR21 | ||||
| Swissprot | Q8H7S7 | 1e-121 | ORR21_ORYSJ; Two-component response regulator ORR21 | ||||
| Swissprot | Q8L9Y3 | 1e-124 | ARR14_ARATH; Two-component response regulator ARR14 | ||||
| TrEMBL | A0A067KT22 | 0.0 | A0A067KT22_JATCU; Two-component response regulator | ||||
| STRING | XP_010553611.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8150 | 27 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01760.1 | 1e-125 | response regulator 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



