 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010554450.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 466aa MW: 51929.6 Da PI: 6.263 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40.1 | 6.6e-13 | 319 | 365 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
XP_010554450.1 319 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLADAITYITDLQ 365
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 3.5E-42 | 48 | 227 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| SuperFamily | SSF47459 | 6.54E-18 | 315 | 381 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.609 | 315 | 364 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 9.87E-15 | 318 | 369 | No hit | No description |
| Pfam | PF00010 | 1.8E-10 | 319 | 365 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 6.7E-17 | 319 | 380 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.6E-15 | 321 | 370 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 466 aa Download sequence Send to blast |
MGHKFWENQE DRAVVESIIG SEACEFLISS ASNSSLSKLV VPPNDPNLQQ GLRHVVEGSD 60 WDYAIFWLAS SLSNSDGCLL IWGDGHCRAP KGKRGISNDV DPKQEEVKRL ALQKLHMSFG 120 GSDHDNSMAQ LEGVTDLEMF FLASMYFSFR CESAKYGPAG IYTTGKPLWA ADLPSCLNYY 180 RVRSFLSRSA GFETVLSVPV KSGVVELGSF RTIPEDKSVI EMVKAVFGAA DPIQTKAAPK 240 IFGRELSLGG SKPQTMSINF SPKMEDDSGF SLESYDAIGG SNQVYGYEQG KYDVLCLTDE 300 QKPRKRGRKP ANGREEALNH VEAERQRREK LNQRFYALRA VVPNISKMDK ASLLADAITY 360 ITDLQKKIRV FETEKQIMNR REINQTPPAE IDFQQRQDDA VVRLSCPLET HPVSKVIQAF 420 RENQVTAHDS NVTATEDCMV HTFSVRAQDG VTAEQLKEKL AASFSQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| 5gnj_B | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| 5gnj_E | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| 5gnj_F | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| 5gnj_G | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| 5gnj_I | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| 5gnj_M | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| 5gnj_N | 8e-27 | 313 | 382 | 2 | 72 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
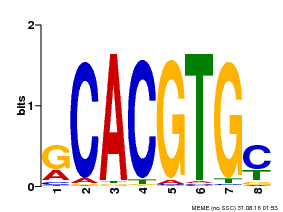
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010554450.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010554450.1 | 0.0 | PREDICTED: transcription factor bHLH3 | ||||
| Swissprot | O23487 | 0.0 | BH003_ARATH; Transcription factor bHLH3 | ||||
| TrEMBL | A0A178V653 | 0.0 | A0A178V653_ARATH; JAM3 | ||||
| STRING | XP_010554450.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6611 | 27 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 0.0 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



