 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010556598.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 593aa MW: 65507.5 Da PI: 7.1974 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40 | 7.2e-13 | 436 | 481 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr+++P+ + K++Ka+ L Av YI++L
XP_010556598.1 436 NHVEAERQRREKLNQRFYALRTVVPNiS------KMDKASLLGDAVAYINEL 481
799***********************66......***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 2.9E-51 | 48 | 236 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.901 | 432 | 481 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.25E-14 | 435 | 486 | No hit | No description |
| SuperFamily | SSF47459 | 1.57E-17 | 435 | 496 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.9E-10 | 436 | 481 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.9E-17 | 436 | 493 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.1E-15 | 438 | 487 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 593 aa Download sequence Send to blast |
MNIGCVVWNE DDKDIVGSLL GKRGLDYLLS NSVSNENHLM TMASDENLQD KLSDLVERPN 60 ASNFSWNYAI FWQISRSKTG DSVLCWGDGS CREPKEGEES EIIRILSIGR EEETQQRIRK 120 RVLQKLHTFF GGSEEENYAF GLDRVTDTEM FFLASMYFSF PRGEGGPGKC FVSGKPVWLS 180 DVLKSCSDYC VRSFLAKSAG IRTVVLVPTD VGVVELGSMR SLPESHESTQ SIRSLFSSKL 240 THQMVPAIFP QKKDENRILG IGLSNLGVLE RVGGAPKNSG KDLHNLGHIQ HGHNHNQQRQ 300 FREKLAIRKT DDRVPRTPEA YPNGGKLMFS NPPTNTVLSP SWAQTDIYGS LAVSSTPTTA 360 AQATNGKDVP SGDDFRFLPL QPQRPAQIQI DFSGASSRAS DNSDGEGTEW PDIGDDGSRP 420 RKRGRKPANG RAEALNHVEA ERQRREKLNQ RFYALRTVVP NISKMDKASL LGDAVAYINE 480 LHAKLKVMEA ERETSGYSSK PPISREAKIN VQASGEDVTV RVNCPLDSHP ASGILHAFEE 540 SQATVIDSKL EVGQDTVLHT FVIKPGELTK EKLVSAFSRE PSSSLQSRSS SGR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 3e-25 | 430 | 492 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
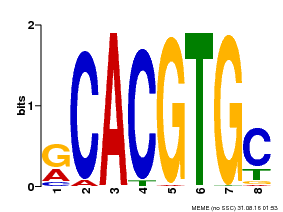
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010556598.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010556598.1 | 0.0 | PREDICTED: transcription factor bHLH13-like | ||||
| Swissprot | Q9LNJ5 | 0.0 | BH013_ARATH; Transcription factor bHLH13 | ||||
| TrEMBL | A0A178WJW5 | 0.0 | A0A178WJW5_ARATH; JAM2 | ||||
| STRING | XP_010556598.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 0.0 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



