 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010558266.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 469aa MW: 52022.2 Da PI: 9.8152 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.3 | 1.3e-05 | 27 | 49 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C++C+k F r nL+ H r H
XP_010558266.1 27 FVCEICNKGFQRDQNLQLHRRGH 49
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 14 | 0.00014 | 104 | 126 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC++C+k + +s+ k H +t+
XP_010558266.1 104 WKCEKCSKKYAVQSDWKAHTKTC 126
58*******************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 1.5E-5 | 26 | 49 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.08E-7 | 26 | 49 | No hit | No description |
| PROSITE profile | PS50157 | 10.949 | 27 | 49 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF12171 | 7.4E-5 | 27 | 49 | IPR022755 | Zinc finger, double-stranded RNA binding |
| SMART | SM00355 | 0.017 | 27 | 49 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 29 | 49 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 67 | 69 | 99 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 4.1E-5 | 92 | 125 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.08E-7 | 99 | 124 | No hit | No description |
| SMART | SM00355 | 130 | 104 | 124 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0045604 | Biological Process | regulation of epidermal cell differentiation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 469 aa Download sequence Send to blast |
MRCAYSTYPD AEVMALSPKT LMATNRFVCE ICNKGFQRDQ NLQLHRRGHN LPWKLRQRTN 60 KEAVKKKVYV CPIKTCVHHE PSRALGDLTG IKKHYSRKHG EKKWKCEKCS KKYAVQSDWK 120 AHTKTCGTRE YKCDCGTLFS RKDSFITHRA FCDALTDETA RITSLGNDNS VSANNLNFGN 180 DNSDNNLNQL PHGFVNRDTV HHPDIDSAIN HHHHQFGLGF GQDLCGMHTQ GLSEMVQMTS 240 TGGNHHLFPS STSSLPDFAG QSHHHHFQIP ATPLNPNLKS SSPMASASLP SLQQHQTLKD 300 TSLSPLFSSS DTQHQNNNKP ISPMPATALL QKAAQMGSTR STASAAPSFF AGSMMSSSAA 360 SPSSSPSPRN PIMIHHQMFN PNPRSINHQE NHNLSNPSPP QKNSATSSST AAPKMANPNP 420 NPGQNSIQQM GMTRDFLGVR SEHHHHHQQQ QQMGRPFLPQ ELARFAPFG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 3e-39 | 100 | 169 | 3 | 72 | Zinc finger protein JACKDAW |
| 5b3h_F | 3e-39 | 100 | 169 | 3 | 72 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that, together with BIB, regulates tissue boundaries and asymmetric cell division by a rapid up-regulation of 'SCARECROW' (SCR), thus controlling the nuclear localization of 'SHORT-ROOT' (SHR) and restricting its action (PubMed:17785527, PubMed:25829440). Binds DNA via its zinc fingers (PubMed:28211915, PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). Confines CYCD6 expression to the cortex-endodermis initial/daughter (CEI/CEID) tissues (PubMed:25829440). Required for radial patterning and stem cell maintenance (PubMed:17785527). Counteracted by 'MAGPIE' (MGP) (PubMed:17785527). Binds to the SCR and MGP promoter sequences (PubMed:21935722). Controls position-dependent signals that regulate epidermal-cell-type patterning (PubMed:20356954). {ECO:0000269|PubMed:17785527, ECO:0000269|PubMed:20356954, ECO:0000269|PubMed:21935722, ECO:0000269|PubMed:24821766, ECO:0000269|PubMed:25829440, ECO:0000269|PubMed:28211915}. | |||||
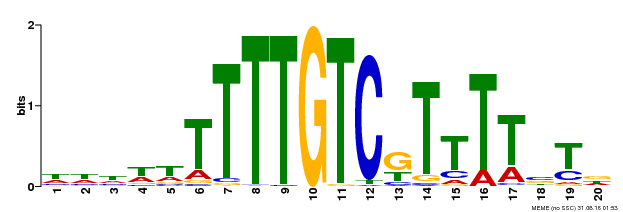
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00485 | DAP | Transfer from AT5G03150 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010558266.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not regulated by SCR and SHR. {ECO:0000269|PubMed:21935722}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010558266.1 | 0.0 | PREDICTED: zinc finger protein JACKDAW-like isoform X2 | ||||
| Swissprot | Q700D2 | 0.0 | IDD10_ARATH; Zinc finger protein JACKDAW | ||||
| TrEMBL | A0A3P6DG06 | 0.0 | A0A3P6DG06_BRACM; Uncharacterized protein | ||||
| STRING | XP_010558265.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G03150.1 | 1e-152 | C2H2-like zinc finger protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



