 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010558628.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 482aa MW: 53559.8 Da PI: 4.7475 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 112.5 | 2.9e-35 | 22 | 113 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkellek 96
Fl+k+y++++d++++e++sws+ +nsfvv + ef+k++LpkyFkh+nf+SFvRQLn+YgF+kv+ + weF+++ F kg+k+ll++
XP_010558628.1 22 FLSKTYDMVDDPSTNEVVSWSSGNNSFVVRNVPEFTKELLPKYFKHNNFSSFVRQLNTYGFRKVDPDC---------WEFANEGFLKGQKQLLKS 107
9***************************************************************9987.........****************** PP
XXXXXX CS
HSF_DNA-bind 97 ikrkks 102
i r+k
XP_010558628.1 108 IVRRKP 113
***986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 7.6E-38 | 16 | 106 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 5.58E-34 | 17 | 111 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 5.6E-59 | 18 | 111 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 4.6E-20 | 22 | 45 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 5.7E-31 | 22 | 111 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 4.6E-20 | 60 | 72 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 61 | 85 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 4.6E-20 | 73 | 85 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009408 | Biological Process | response to heat | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 482 aa Download sequence Send to blast |
MESVRESVPA SVPPAVNSVP PFLSKTYDMV DDPSTNEVVS WSSGNNSFVV RNVPEFTKEL 60 LPKYFKHNNF SSFVRQLNTY GFRKVDPDCW EFANEGFLKG QKQLLKSIVR RKPAHVHNQQ 120 QPQVQNSSVG ACVEVGKFGL EEEVDRLKRD KNVLMQELVR LRQQQQATEH QLQNVGQKVQ 180 TMEQRQQQMM SFLAKAVQSP VFLNQLVQQQ SDGNRCIGGS NKKRRLPGED QEISSSHGAN 240 GLNGQMVKYQ PSMNEAGKAM LQQILKMSSA PKYDPFTNNA GSFLLDDIPS PNALDNGSSN 300 RVSEVTLAEV SPNPSFSCLS SAPTTNLQEP CKTDEVPKTN TSPQSRAETV QPKLAQKQRG 360 AAEESPDSDL VRSCGVDNGE SNLDPITAVL DGFMELEGDV DDMDEFLDGV PKLPVVHDSF 420 WEQFFAASPV TGDTDEIISG AMESIWVMEQ ESQPDQRNGG KNQQMDNLTE QMGLLRSEAQ 480 RK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 2e-25 | 9 | 111 | 18 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 2e-25 | 9 | 111 | 18 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 2e-25 | 9 | 111 | 18 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00514 | DAP | Transfer from AT5G16820 | Download |
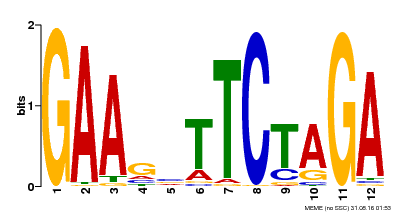
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010558628.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: DNA-binding capacity is reduced by HSBP in vitro. {ECO:0000269|PubMed:20388662}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010558628.1 | 0.0 | PREDICTED: heat stress transcription factor A-1e-like isoform X1 | ||||
| Refseq | XP_010558629.1 | 0.0 | PREDICTED: heat stress transcription factor A-1e-like isoform X1 | ||||
| Swissprot | O81821 | 0.0 | HFA1B_ARATH; Heat stress transcription factor A-1b | ||||
| TrEMBL | V4N7P1 | 0.0 | V4N7P1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_010558628.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2236 | 23 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G16820.2 | 0.0 | heat shock factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



