 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010559169.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 971aa MW: 108018 Da PI: 6.2465 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 133.9 | 5e-42 | 152 | 229 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C adls++k+yhrrhkvCe+hska+++lv g +qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
XP_010559169.1 152 VCQVENCGADLSKVKDYHRRHKVCEMHSKASSALVGGFMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 229
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.1E-33 | 147 | 214 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.539 | 150 | 227 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.88E-39 | 151 | 231 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.1E-30 | 153 | 226 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 5.0E-7 | 747 | 853 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.98E-8 | 747 | 850 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.90E-9 | 749 | 850 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 971 aa Download sequence Send to blast |
MEDRTEGDAH RFYGVSAMDL RAVGERNVEW DSSDWKWDGD LFVARQLNPG SSETMGRQFF 60 PLCSAIPTTG NSSNSSSTWS DEGNIEMMER GTRELDKKRR AVVIEDNNLR DDAGSLTLKL 120 CGHGYPAGDR ESLDINGAKK TKTGGSVPSR AVCQVENCGA DLSKVKDYHR RHKVCEMHSK 180 ASSALVGGFM QRFCQQCSRF HVLQEFDEGK RSCRRRLAGH NKRRRKTNPD TGGNVNPLGD 240 DQTSSYLLIS LLKVLSNMRS NGSDHAGDQD LLSHLLKSLV SHAGEQIGKN IAGLLQGRVQ 300 SSLNVGNSHL LSNGQSPQED LKPLSVNESE MPRQDLYVDG TRGAEGMNCS PKRVRMNDFD 360 LNDIYIDSDD GTEDVERSPP VNPTTSSLDY PLWIRQESHQ SSPPQTSRNS DSASDHSPSS 420 SSIDAQGRTD RIVFKLFGKE PNDFPVVLRG QILDWLSHSP TDMESYIRPG CIVLTIYLRQ 480 AEGAWEELCS DLGFSLRRLL DLSDDPFWTD GWIYVRVQNQ IAFALNGQIV VDTSLPLRSR 540 NYSQIFCVRP IAVAATGRVQ FTVRGTNLRK PGTRLLCAVE GKYLVQEATV ESMVGNDALE 600 EESNEVECIA FSCDMPAASG RGFMEVEDNG LSCSFFPFIV VEDDVCSEIR MLESTLEFTG 660 TDSAKQAMDF IHEIGWLLHR SKLNSRLGQE SKPNGNLFPL RRFKWLIEFS MDHEWCAVIR 720 KLLTILFDGT VGSELGPSSD SALSELCLLH RAVRKNSRPM VDMLLRYDPK EQKLLGNEIR 780 MFRPDATGPA GLTPLHIAAG KDGSEDVLDA LTEDPGMVGL EAWKNSRDST GFTPEDYARL 840 RGHFSYIHLI QRKLKKKSTT EEHAVVVDIP LSFPDTAMEK SHQKQKTGLR PSGLEITLAN 900 QAKSSHCKLC DQKMAYGFGP AVRSVAYRPA MLSMVAIAAV CVCVALLFKS CPEVLYVFQP 960 FRWELLAYGS S |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 8e-32 | 152 | 226 | 10 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
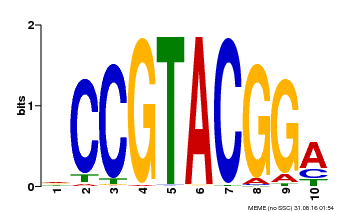
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010559169.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010559169.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A1J3IQZ6 | 0.0 | A0A1J3IQZ6_NOCCA; Squamosa promoter-binding-like protein 1 | ||||
| STRING | XP_010559169.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



