 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010904556.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 601aa MW: 64047.1 Da PI: 9.1087 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.1 | 3.1e-05 | 75 | 97 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
XP_010904556.1 75 FICEVCNKGFQREQNLQLHRRGH 97
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 11.8 | 0.00073 | 151 | 173 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC +C+k + +s+ k H +t+
XP_010904556.1 151 WKCDKCSKKYAVQSDWKAHSKTC 173
58*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 6.7E-6 | 74 | 97 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 6.84E-7 | 74 | 97 | No hit | No description |
| Pfam | PF12171 | 3.0E-5 | 75 | 97 | IPR022755 | Zinc finger, double-stranded RNA binding |
| PROSITE profile | PS50157 | 10.99 | 75 | 97 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0052 | 75 | 97 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 77 | 97 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 230 | 116 | 146 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 6.5E-4 | 139 | 172 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 6.84E-7 | 146 | 171 | No hit | No description |
| SMART | SM00355 | 150 | 151 | 171 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 601 aa Download sequence Send to blast |
MASASSTEPF FGIREEDHRN QMKQQQPASM PTSSSAAAPQ TAAPVKKKRN LPGNPNPDAE 60 VIALSPKALL ATNRFICEVC NKGFQREQNL QLHRRGHNLP WKLRQKSTKE VRRRVYVCPE 120 TTCVHHDPSR ALGDLTGIKK HFCRKHGEKK WKCDKCSKKY AVQSDWKAHS KTCGTREYRC 180 DCGTLFSRRD SFITHRAFCD ALAQESARIP TGLSTIGSHL YGNSSMALGL SQVNSQITSS 240 LQVQSHQSSN NILRLGGGSG SGSHFEHLIS SSNPSPFRPP QPLPSSAFFL GGASNQDFNE 300 ETQSHHSLLQ NKPFHGLMQL PDLQSRATAS SSTAAAANLF NLGFFSNSSS TSSISTSNIT 360 SNQNNHLLIS DPFSSGNGST ESTTLFSGNF MSDHMAAGMS PLLNTSMRNE SSLPQMSATA 420 LLQKAAQMGS TTSGGSSLLQ GFSSASPSGS RSTSFQAIFN GGSNGSGDAL RFQMENETRL 480 QDLMNSLANG NAGLFGGSGG VAAFAGSCTT GGHEQQAGGI GGFNSGLCNM DEAKFQHNIS 540 PGNLGGSDGL TRDFLGVGNM VRSMGGGLSQ RGRHHGIDIS SMDHEMKSAT SSQSFDGGTL 600 Q |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 7e-34 | 147 | 211 | 3 | 67 | Zinc finger protein JACKDAW |
| 5b3h_F | 7e-34 | 147 | 211 | 3 | 67 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a positive regulator of the starch synthase SS4. Controls chloroplast development and starch granule formation (PubMed:22898356). Binds DNA via its zinc fingers (PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). {ECO:0000269|PubMed:22898356, ECO:0000269|PubMed:24821766}. | |||||
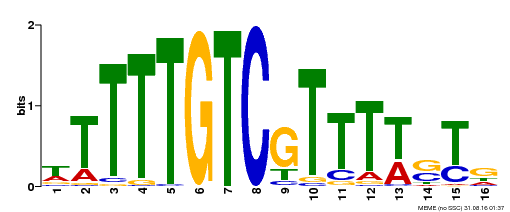
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010904556.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Refseq | XP_010904558.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Refseq | XP_010904560.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Refseq | XP_029116694.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Swissprot | Q9ZUL3 | 1e-131 | IDD5_ARATH; Protein indeterminate-domain 5, chloroplastic | ||||
| TrEMBL | A0A2H3ZAW8 | 0.0 | A0A2H3ZAW8_PHODC; protein indeterminate-domain 5, chloroplastic-like | ||||
| STRING | XP_008811073.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1733 | 37 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02070.1 | 1e-101 | indeterminate(ID)-domain 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



