 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010905300.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 672aa MW: 72492.3 Da PI: 6.5093 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 100.3 | 1.2e-31 | 264 | 321 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C++kk++ers+ d++++ei+Y+g+H+h+k
XP_010905300.1 264 EDGYNWRKYGQKHVKGSEYPRSYYKCTHPNCQMKKHLERSH-DGQITEIIYKGRHDHPK 321
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 33.8 | 6.6e-11 | 439 | 460 | 1 | 22 |
---SS-EEEEEEE--TT-SS-E CS
WRKY 1 ldDgynWrKYGqKevkgsefpr 22
ldDgy+WrKYGqK+vkg+++pr
XP_010905300.1 439 LDDGYRWRKYGQKVVKGNPNPR 460
59*******************9 PP
| |||||||
| 3 | WRKY | 52.7 | 8.7e-17 | 484 | 520 | 23 | 59 |
EEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 23 sYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
sYY+Ct agCpv+k+ver+++dpk+v++tYeg+Hnh+
XP_010905300.1 484 SYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHD 520
8***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.8E-27 | 256 | 322 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.09E-24 | 262 | 322 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.927 | 263 | 322 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.2E-34 | 263 | 321 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.3E-23 | 264 | 320 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-33 | 424 | 522 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.22E-21 | 431 | 522 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.305 | 434 | 522 | IPR003657 | WRKY domain |
| SMART | SM00774 | 9.9E-35 | 439 | 521 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.6E-7 | 440 | 461 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.7E-12 | 484 | 520 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 672 aa Download sequence Send to blast |
MAEDPEADAG DLHVGAPLEG SRGPELGLDG FGEGNGGGSW EGRGGREGRA DPRVSSLAGA 60 RYKSMSPARL PIARSPCLTI SSGLSPSALL ESPVLLTNMK AEPSPTTGTF SMPSLMNKAV 120 CSDTISSPRD SNSNACDEES SAAFEFKRHM RSSMNSGLSP LGALASVNPT QQEQEPSLPV 180 QSQSQTQTSS QIAKAEKATL SSDELTLSVR NPHPPLETLP SAGVPPEVAS DELQQIKGSE 240 NGIQSLQSDH NGSMPSTIIE KSLEDGYNWR KYGQKHVKGS EYPRSYYKCT HPNCQMKKHL 300 ERSHDGQITE IIYKGRHDHP KPQPSRRVAV GALLCTQGEE KPEGLSSLAS PEGKSSTVHI 360 SYPIDPTGAP QLSPISASDD DVEIGGARSH YTGDEVTDGD DPESKRRKMD ATSVDATPIG 420 KPNREPRVVV QTLSEVDILD DGYRWRKYGQ KVVKGNPNPR YCKMRHLSPL RSQICRLRDI 480 KYGSYYKCTN AGCPVRKHVE RASHDPKAVI TTYEGKHNHD VPAAKTVNHD TSVPTIADGS 540 NTMMTHTSSA LRGFMRNGDT MIISQQYTQP EESDTISLDL GVGISRNHSN TNELQQSPAR 600 DQVHGHQIQS VGSDCSNAMI QANPLSTLYG NSSNGVYGSS EDKSDGFIFR AAPMDCSSNL 660 YYASTGSLVM GP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-32 | 263 | 523 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-32 | 263 | 523 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
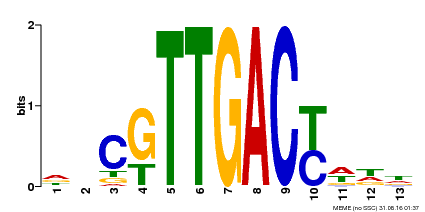
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010905301.1 | 0.0 | WRKY transcription factor SUSIBA2 isoform X1 | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | A0A2H3YMB2 | 0.0 | A0A2H3YMB2_PHODC; WRKY transcription factor SUSIBA2-like isoform X1 | ||||
| STRING | XP_008800638.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6740 | 38 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 1e-136 | WRKY family protein | ||||



