 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010907513.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 473aa MW: 52109.9 Da PI: 7.4514 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 55.1 | 1.9e-17 | 160 | 209 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta+ Aa+a+++a+ k++g
XP_010907513.1 160 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAQAAARAYDRAAIKFRG 209
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 42.1 | 2.1e-13 | 252 | 302 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ + +k+++lg f+++ eAa+a+++a+ k +g
XP_010907513.1 252 SKYRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDSEIEAARAYDKAAIKCNG 302
89********.7******5553..2.26**********99**********99776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 2.3E-9 | 160 | 209 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.19E-16 | 160 | 217 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 4.09E-11 | 161 | 216 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 3.3E-17 | 161 | 217 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.886 | 161 | 217 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.7E-31 | 161 | 223 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.1E-9 | 252 | 302 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 6.46E-25 | 252 | 312 | No hit | No description |
| SuperFamily | SSF54171 | 8.5E-17 | 252 | 311 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 15.712 | 253 | 310 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.3E-15 | 253 | 310 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.0E-31 | 253 | 316 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.5E-6 | 254 | 265 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.5E-6 | 292 | 312 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 473 aa Download sequence Send to blast |
MMMLDLNLAS SGESLAFQEK PLPDEDSGTS NSSVLNAEAS SNAPDDDSCS TRPTAGFQFG 60 ILRVSASAEG ENEMEEEIKG RVNNVSLESN FLTRQLFPPA SPAVPEWPQL PAVAASSSAS 120 SRQHLLDLRF SQSGNPADVR VLQQQQHVKK SRRGPRSRSS QYRGVTFYRR TGRWESHIWD 180 CGKQVYLGGF DTAQAAARAY DRAAIKFRGV DADINFNLVD YEEDLKQTRN LTKEEFVHIL 240 RRQSTGFSRG SSKYRGVTLH KCGRWEARMG QFLGKKYIYL GLFDSEIEAA RAYDKAAIKC 300 NGRDAVTNFE PSTYEGELLT EANSEATGHD VDLNLRISQP VAHSPKKDHN SIGIQFHYGL 360 LESSDAKKVT IDSTSSQLAG QPHHVWTAQR PALFPTIEEG AREKRLEVGS QALPAWAWQM 420 HGPTPLPLFS SAASSGFSTT AVTSAHPSSL PPPSATLHSQ FSLPAPSNFR FRS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition. Regulator of starch biosynthesis especially during seed development (e.g. endosperm starch granules); represses the expression of type I starch synthesis genes. Prevents lemma and palea elongation as well as grain growth (By similarity). {ECO:0000250|UniProtKB:P47927, ECO:0000250|UniProtKB:Q2TQ34}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
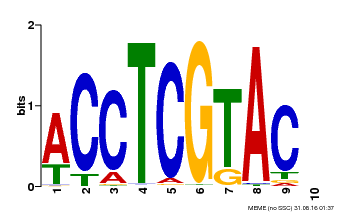
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010907513.1 | 0.0 | APETALA2-like protein 1 isoform X1 | ||||
| Swissprot | B8AXC3 | 1e-138 | AP21_ORYSI; APETALA2-like protein 1 | ||||
| TrEMBL | A0A2H3ZP21 | 0.0 | A0A2H3ZP21_PHODC; AP2-like ethylene-responsive transcription factor TOE3 isoform X1 | ||||
| STRING | XP_008789029.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2448 | 38 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-110 | related to AP2.7 | ||||



