 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010907773.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 592aa MW: 66314.6 Da PI: 4.8933 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 80.7 | 1.7e-25 | 200 | 253 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++W+ +LH++Fv+av+q+ G +k Pk+il+lm+++gLt+e+v+SHLQkYRl
XP_010907773.1 200 KARVVWSVDLHQKFVNAVNQI-GFDKVGPKKILDLMNIPGLTRENVASHLQKYRL 253
68*******************.9*******************************8 PP
| |||||||
| 2 | Response_reg | 78.6 | 2.2e-26 | 19 | 127 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaG 93
vl+vdD+p+ +++l+++l+k y ev+++ ++ ale+l+e++ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + + ++ G
XP_010907773.1 19 VLVVDDDPTWLKILEKMLRKCSY-EVTTCGLARVALEMLRERKdhFDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSIDGETSRVMKGVQHG 111
89*********************.***************777777**********************6644.8********************** PP
ESEEEESS--HHHHHH CS
Response_reg 94 akdflsKpfdpeelvk 109
a d+l Kp+ ++el++
XP_010907773.1 112 ACDYLLKPVRMKELRN 127
*************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 6.4E-152 | 2 | 582 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 1.57E-34 | 16 | 139 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 1.0E-43 | 16 | 153 | No hit | No description |
| SMART | SM00448 | 2.2E-31 | 17 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.603 | 18 | 133 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.1E-23 | 19 | 128 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 5.92E-29 | 20 | 132 | No hit | No description |
| SuperFamily | SSF46689 | 6.09E-18 | 197 | 257 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-27 | 199 | 258 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.3E-22 | 200 | 253 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.3E-8 | 202 | 252 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 592 aa Download sequence Send to blast |
MANPPASTRT EAFPVGLRVL VVDDDPTWLK ILEKMLRKCS YEVTTCGLAR VALEMLRERK 60 DHFDIVISDV NMPDMDGFKL LEHVGLEMDL PVIMMSIDGE TSRVMKGVQH GACDYLLKPV 120 RMKELRNIWQ HVYRKKMHEV KEIEGHYINE DIQIMKIGSD DFEDRHVSSG ADINYVRKRK 180 DVDNKEHSDQ EFSDPATVKK ARVVWSVDLH QKFVNAVNQI GFDKVGPKKI LDLMNIPGLT 240 RENVASHLQK YRLYLSRLQK QNEGRTSSGN LQSDFSSKDV AGNFDVLSST QMQPSGVSSK 300 FICTGQKIQA QGIIPDDHAD DFKSSLSLQV TGSKQALVGD IPDSQGVISV SRLGILLPFK 360 DMIPDAGKKS LEPAISRNQS WSGSIPVMQF MQHPRHDHEC CCLIEAYSCL PKPDQEQVFD 420 HFDTPPSVIS NTCNTENDIS GLLEFKLVHT NHRSSDTKTA SQLSCMTDTT SAQVDSGSSN 480 TQDYGVAFGI DGLTNIKDYY HTQTSSYLTP PSGELSLKNE TDLVSSPEDL QFCSPQGIGS 540 FENVGFNSME IFQYDDSTSV TEVHSNWYDG LEFNSDYLYD SVDPVIDECL FA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-17 | 199 | 258 | 4 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
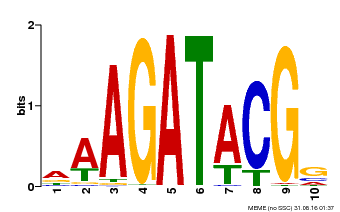
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010907773.1 | 0.0 | two-component response regulator ORR26 isoform X1 | ||||
| Swissprot | Q5N6V8 | 0.0 | ORR26_ORYSJ; Two-component response regulator ORR26 | ||||
| TrEMBL | A0A2H3YDP8 | 0.0 | A0A2H3YDP8_PHODC; Two-component response regulator | ||||
| STRING | XP_008797265.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5900 | 37 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-124 | response regulator 11 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



