 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010908480.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 241aa MW: 25465.8 Da PI: 8.5961 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 141.5 | 2.7e-44 | 26 | 125 | 1 | 100 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkaela 95
+CaaCk+lrrkC++dCv+apyfp +qp+kf vh++FGasnv+kll++l++ +reda++sl+yeA++r+rdPvyG+vgvi+ lq+ql+ql+ +l+
XP_010908480.1 26 PCAACKFLRRKCQPDCVFAPYFPPDQPQKFVHVHRVFGASNVTKLLNELHPYQREDAVNSLAYEADMRLRDPVYGCVGVISILQHQLRQLQMDLS 120
7********************************************************************************************** PP
DUF260 96 llkee 100
+k+e
XP_010908480.1 121 CAKSE 125
99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 26.899 | 25 | 126 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 7.1E-44 | 26 | 123 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009799 | Biological Process | specification of symmetry | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0009954 | Biological Process | proximal/distal pattern formation | ||||
| GO:0048441 | Biological Process | petal development | ||||
| GO:0005654 | Cellular Component | nucleoplasm | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 241 aa Download sequence Send to blast |
MASSSAPVPA SSAAVSSSMT ASTSSPCAAC KFLRRKCQPD CVFAPYFPPD QPQKFVHVHR 60 VFGASNVTKL LNELHPYQRE DAVNSLAYEA DMRLRDPVYG CVGVISILQH QLRQLQMDLS 120 CAKSELSKYQ SAAGSSAPTF GGGLATAGVH GFLNADHPIG FGREQFFPAG NNREPPRPNQ 180 HLMMRNFEGE LVGRLGANGA YDAAGIVAAM NASAASIGPL GGQFLKPSSA GRNDRPSVGP 240 S |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 7e-54 | 16 | 139 | 1 | 124 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 7e-54 | 16 | 139 | 1 | 124 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Promotes the switch from proliferation to differentiation in the embryo sac. Negative regulator of cell proliferation in the adaxial side of leaves. Regulates the formation of a symmetric lamina and the establishment of venation. Interacts directly with RS2 (rough sheath 2) to repress some knox homeobox genes. {ECO:0000269|PubMed:17209126}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00211 | DAP | Transfer from AT1G65620 | Download |
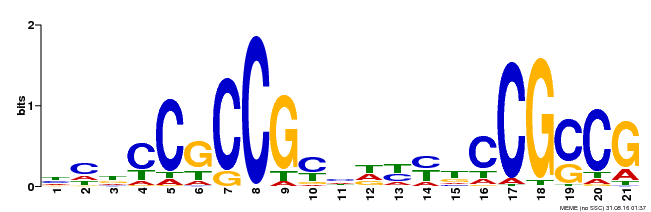
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_029118870.1 | 1e-152 | protein ASYMMETRIC LEAVES 2 | ||||
| Swissprot | Q32SG3 | 2e-80 | LBD6_MAIZE; LOB domain-containing protein 6 | ||||
| TrEMBL | A0A2H3ZNL3 | 1e-130 | A0A2H3ZNL3_PHODC; LOB domain-containing protein 6-like | ||||
| STRING | XP_008788266.1 | 1e-131 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2267 | 38 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G65620.4 | 2e-58 | LBD family protein | ||||



