 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010918103.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 281aa MW: 30879.9 Da PI: 9.5451 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 106 | 2.1e-33 | 27 | 81 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWtp+LH+rFv+av++LGG+ekAtPk++l++m++kgLtl+h+kSHLQkYRl
XP_010918103.1 27 KPRLRWTPDLHDRFVDAVAKLGGPEKATPKSVLRVMGMKGLTLYHLKSHLQKYRL 81
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.97 | 24 | 84 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.9E-32 | 24 | 82 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 7.88E-16 | 27 | 83 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-24 | 27 | 82 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.4E-9 | 29 | 80 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.8E-24 | 122 | 169 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 281 aa Download sequence Send to blast |
MERGYGSAYE AAAGGGGGGV VLSRDPKPRL RWTPDLHDRF VDAVAKLGGP EKATPKSVLR 60 VMGMKGLTLY HLKSHLQKYR LGKQSRKETG VESKKDGSNS SGITYSSATT GSLPRGNNAG 120 EMPLAEALKY QLEVQRKLHE QLEVQKKLQM RIEAQGKYLQ AILEKAQKSL SFDMNNSGSL 180 EATRAQLTDF NLALSGLMDN VNQVCEEKNE ELGQFIPQDN LKRAHSSGFQ LYQKQQEDGE 240 DIKLAPHGGS LLLDLNVKGN YELFGGPRGS ELDLRIHSPR G |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 4e-21 | 27 | 83 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-21 | 27 | 83 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-21 | 27 | 83 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-21 | 27 | 83 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 4e-21 | 27 | 83 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
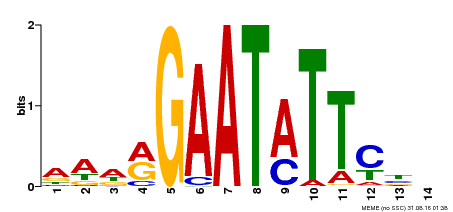
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010918102.1 | 0.0 | myb family transcription factor PHL11 | ||||
| Refseq | XP_010918103.1 | 0.0 | myb family transcription factor PHL11 | ||||
| Swissprot | C0SVS4 | 1e-73 | PHLB_ARATH; Myb family transcription factor PHL11 | ||||
| TrEMBL | A0A2H3YKB2 | 0.0 | A0A2H3YKB2_PHODC; myb family transcription factor PHL11 | ||||
| STRING | XP_008800016.1 | 0.0 | (Phoenix dactylifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 4e-74 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



