 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010922496.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 482aa MW: 52358.7 Da PI: 6.5114 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.5 | 6e-17 | 167 | 216 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
XP_010922496.1 167 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 216
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 43 | 1.1e-13 | 259 | 309 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ + +k+++lg f+++ eAa+a+++a+ k++g
XP_010922496.1 259 SKYRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDSEIEAARAYDKAAIKFNG 309
89********.7******5553..2.26**********99***********9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.11E-15 | 167 | 224 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 5.5E-9 | 167 | 216 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.5E-31 | 168 | 230 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.794 | 168 | 224 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.26E-11 | 168 | 225 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 6.6E-17 | 168 | 224 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.29E-25 | 259 | 319 | No hit | No description |
| SuperFamily | SSF54171 | 7.19E-17 | 259 | 318 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 5.4E-9 | 259 | 309 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 5.7E-32 | 260 | 323 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.291 | 260 | 317 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 6.7E-16 | 260 | 317 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-6 | 261 | 272 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-6 | 299 | 319 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 482 aa Download sequence Send to blast |
MVLDLNVASP GESSSCGEEK VVMEGSGGAP MEESGTSNSS VLNAEASSNA GDDDSCSAAA 60 AVGGFNFGIL KSSLRAHGEE EVEEELEEED GASGEAGFVT RQLFPPFSVV QEGVQPPTVA 120 SSSTPALPRR MELRFLQSNT AGSVEAKVLQ QQHQVKKSRR GPRSRSSQYR GVTFYRRTGR 180 WESHIWDCGK QVYLGGFDTA HAAARAYDRA AIKFRGVDAD INFNICDYEE DLKQMRNLTK 240 EEFVHILRRQ STGFARGSSK YRGVTLHKCG RWEARMGQFL GKKYIYLGLF DSEIEAARAY 300 DKAAIKFNGR EAVTNFEPST YEGEVHPEPN NEVAGDNVDL DLRISQPNVH SPKKDNNSMD 360 IQPNHSSFAA SGAGNSRANN STSHLAHPQH LPMAPEHPHA WTALYPGFFT HTEERATKGS 420 EVGFPAPSNR AWPTQGPTSP FSSAASSGFS TAATVVAAHH LPPSAAHLNF PPSATVNHYH 480 MS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with IDS1, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). Regulates the transition from spikelet meristem to floral meristem, spikelet meristem determinancy and the floral organ development (PubMed:17144896). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:17144896, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
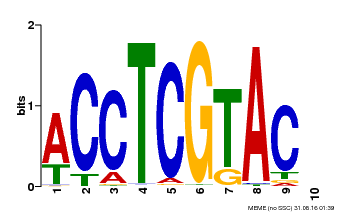
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:20017947, ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010922496.1 | 0.0 | APETALA2-like protein 2 | ||||
| Swissprot | Q8H443 | 1e-123 | AP23_ORYSJ; APETALA2-like protein 3 | ||||
| TrEMBL | A0A2H3ZLG9 | 0.0 | A0A2H3ZLG9_PHODC; LOW QUALITY PROTEIN: AP2-like ethylene-responsive transcription factor TOE3 | ||||
| STRING | XP_008802331.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2448 | 38 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-103 | related to AP2.7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



