 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010923540.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 793aa MW: 86501.4 Da PI: 5.5446 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 51.6 | 1.8e-16 | 488 | 526 | 2 | 40 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
+k+CnCkkskClk+YCeCfaag++Cse C+C++C+Nk
XP_010923540.1 488 ACKRCNCKKSKCLKLYCECFAAGVYCSEPCSCQGCFNKP 526
589**********************************96 PP
| |||||||
| 2 | TCR | 50.5 | 4e-16 | 573 | 611 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
XP_010923540.1 573 RHKRGCNCKKSSCLKKYCECYQGGVGCSISCRCEGCKNA 611
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 8.8E-19 | 487 | 528 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.792 | 488 | 613 | IPR005172 | CRC domain |
| Pfam | PF03638 | 4.9E-12 | 490 | 525 | IPR005172 | CRC domain |
| SMART | SM01114 | 4.4E-17 | 573 | 614 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 1.3E-11 | 575 | 611 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 793 aa Download sequence Send to blast |
MDTPERSKIG GTPLSKFEDS PVFNFINSLS PIQPVKSVDS IHTEQSYLQP VSSIFTSPHA 60 NLQRESRFLI RNSLSEFSKD ESSSDNVNES NLCTGVQNAV SLSRCTVIAQ ENCSVICSLN 120 EATVDPPDEC SILPSNLPQS EQHDSGSPDH NTAPWYDIKM DVTLDIDRAP VELVHFVQNG 180 LDKRKSLLAS ESEFLENYPL EQNKDEAVGC DWVNLISDNA DVPLIFDSNN ESEVCRGPIE 240 ELGDSSKPAQ DLDNSQKTQP DESFGPCVQN AAQDPPVNHS AENGKEDETA HTLQILSGTC 300 QNQVLLSNGT SDQIQSGHKV ESQQQRGIRR RCLVFDMAGI SKKNLQSNSN QHSSSSLLSN 360 EKMASDDKCS TPGTSQAPCV LPGIGLHLNT LATTSKERPV TQEMLASGKE LISMPCPVGP 420 FGPTTSGWKG KSLAVEKDVS PTENEVQNLQ IMHNDASQAP ALGNGEELNQ VSPKKKRRKS 480 EHGGESEACK RCNCKKSKCL KLYCECFAAG VYCSEPCSCQ GCFNKPIHEE TVLATRKQIE 540 SRNPLAFAPK VIRTSETGQE MGEDANKTPA SARHKRGCNC KKSSCLKKYC ECYQGGVGCS 600 ISCRCEGCKN AFGRKEGIVL IGAEEIEQGE HETDHCEREK DKPDDDGQQH ATVQIDENNS 660 SEGLLSVTPL QTCRPSVKLS FCSSAKPPRS STLPVGFSPW LYTSQHFQKS EILPQHKLEK 720 HVNTALEDDT PDVLRCDAPP SNGVKTTSPN RKRVSPPHNG IGLLPSRKGG RKVILKSIPS 780 FPSLTGDASE DYP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 4e-17 | 492 | 610 | 14 | 120 | Protein lin-54 homolog |
| 5fd3_B | 4e-17 | 492 | 610 | 14 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 473 | 477 | KKKRR |
| 2 | 473 | 478 | KKKRRK |
| 3 | 474 | 478 | KKRRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
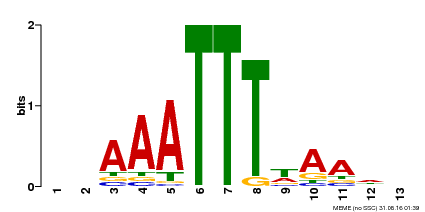
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010923540.1 | 0.0 | protein tesmin/TSO1-like CXC 3 | ||||
| TrEMBL | A0A2H3XX47 | 0.0 | A0A2H3XX47_PHODC; protein tesmin/TSO1-like CXC 2 | ||||
| STRING | XP_008789994.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6147 | 34 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22780.1 | 1e-125 | Tesmin/TSO1-like CXC domain-containing protein | ||||



