 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010924436.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 463aa MW: 51717.7 Da PI: 10.3865 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 58.4 | 1.3e-18 | 8 | 97 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT.---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkv 96
ffkv l+s+ l +p f ++ + +e + t++l+ +sg +W+v++ k+s++ ++++GWk+Fv ++ ++ gDf+vF++dg f v v
XP_010924436.1 8 FFKVY--FPELSSDHLKIPPAFR-KYiE--HELPGTVSLKGPSGGTWNVEF--VKSSKGLLFANGWKKFVADHSIELGDFLVFRYDGGLHF--SVLV 95
55666..34567778******97.5525..458889***************..*******************************9985555..6666 PP
E- CS
B3 97 fr 98
++
XP_010924436.1 96 LD 97
55 PP
| |||||||
| 2 | B3 | 55.8 | 8.3e-18 | 359 | 447 | 4 | 98 |
E-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 4 vltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
v+ s+v s + +p +f++eh ++ sk++tl d++g+ W v++ +s+r +++GW +F an+L+ D++vF+l++r+ +v++fr
XP_010924436.1 359 VMRDSYVYVSFFMNIPYPFVREHL--PKISKKMTLWDPNGKPWAVNY--VSYSSRGGFSAGWGGFSYANNLEKYDVCVFELIKRDHM--KVHIFR 447
567789999999***********5..347899***************..6666666699**********************986555..999998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 2.94E-25 | 2 | 101 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 8.3E-23 | 3 | 101 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 8.50E-20 | 6 | 97 | No hit | No description |
| PROSITE profile | PS50863 | 14.311 | 6 | 99 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 9.2E-16 | 8 | 95 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 5.8E-15 | 8 | 99 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02178 | 1.8 | 261 | 269 | IPR017956 | AT hook, DNA-binding motif |
| Pfam | PF02178 | 5.5 | 282 | 291 | IPR017956 | AT hook, DNA-binding motif |
| Gene3D | G3DSA:2.40.330.10 | 2.5E-25 | 347 | 447 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 6.67E-26 | 349 | 447 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 6.49E-24 | 354 | 447 | No hit | No description |
| SMART | SM01019 | 3.7E-15 | 355 | 449 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.197 | 356 | 449 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.2E-15 | 359 | 447 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0034613 | Biological Process | cellular protein localization | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 463 aa Download sequence Send to blast |
MERSSPHFFK VYFPELSSDH LKIPPAFRKY IEHELPGTVS LKGPSGGTWN VEFVKSSKGL 60 LFANGWKKFV ADHSIELGDF LVFRYDGGLH FSVLVLDATA CEKEAAFLAR PYGGGHIAIV 120 DGIEGGEEGE TMGEHDSKAL VLSPNESFGV GHMRNICSVD CLAMQGPPSK TEKRKKGSPS 180 DEVAAISQDD LDSWKLRKSK SKTPDWSNSM VVARISPPSG ALAVPHKKVH ENTMREKDLH 240 KRKIVSSEHT MAVNKAKTKL QRGRPAKTSA LSVTSTAKSL SKKVGRPSKM SALKQSSQMS 300 ERLKLLPCND VARRGEVVAK VQRMPSLISQ RRPVTKEEIN KALEQAKSFK SKSPFVRVVM 360 RDSYVYVSFF MNIPYPFVRE HLPKISKKMT LWDPNGKPWA VNYVSYSSRG GFSAGWGGFS 420 YANNLEKYDV CVFELIKRDH MKVHIFRVVK EIAPLIRNTK CKS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4i1k_A | 5e-19 | 326 | 448 | 19 | 142 | B3 domain-containing transcription factor VRN1 |
| 4i1k_B | 5e-19 | 326 | 448 | 19 | 142 | B3 domain-containing transcription factor VRN1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00192 | DAP | Transfer from AT1G49480 | Download |
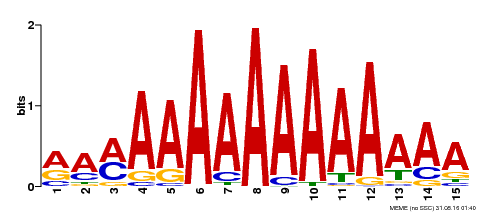
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010924436.1 | 0.0 | B3 domain-containing protein Os11g0197600 isoform X1 | ||||
| TrEMBL | A0A2H3XRJ4 | 0.0 | A0A2H3XRJ4_PHODC; B3 domain-containing protein Os11g0197600-like isoform X1 | ||||
| STRING | XP_008787667.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4461 | 37 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49480.1 | 3e-19 | related to vernalization1 1 | ||||



