 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010926185.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 588aa MW: 62799.6 Da PI: 6.6283 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 97 | 1.2e-30 | 330 | 388 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv+k+v+r+aed +v+++tYeg+Hnh+
XP_010926185.1 330 TDGCQWRKYGQKMAKGNPCPRAYYRCTMAaGCPVRKQVQRCAEDRSVLITTYEGNHNHP 388
7***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.4E-33 | 314 | 390 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 8.5E-28 | 322 | 390 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.815 | 324 | 390 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.7E-37 | 329 | 389 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.5E-27 | 331 | 388 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0016036 | Biological Process | cellular response to phosphate starvation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0080169 | Biological Process | cellular response to boron-containing substance deprivation | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 588 aa Download sequence Send to blast |
MERMDKGGGG LSLGDSAGFH NGGGMDSFLG GGLKQKNSSM ESGHHFQMSL GFHNEPEGRE 60 NRRVVGEMDF FSDDRKERAR PALDHNAPNL CIKKEDLTIN MGLHLLTTNT RSDQSTVDDG 120 LSPNEEDKES KSELAAMQAE LGRVNQENQR LKGMLGHVTN NYNSLQMQFI TLMQERSRRN 180 GSPQSHEVSG DQMGDKRNEH EDAMVPRQFL DLGPASLAED ASHSATEGGS RDRSTSPPNN 240 MEVMPIDNSL KKNSNGKEIA TLDCERTDVR DIRPTFGEVS PDQVSQGWVP NKAPKLSPPK 300 SNTSDQQAQE ATMRKARVSV RARSEAPMIT DGCQWRKYGQ KMAKGNPCPR AYYRCTMAAG 360 CPVRKQVQRC AEDRSVLITT YEGNHNHPLP PAAMAMASTT SAAANMLLSG SMSSTDGLMN 420 SNFLARTILP CSSSMATISA SAPFPTVTLD LTHTPNAQFQ RTSTQFQAPF PNGAPGFGST 480 PPTSLPQVFG QTLFNQSKFS GVQMSAGDGA QFSHPKPQSM QPSTSLADTV SAATAAITAD 540 PNFTAALAAA ITSIIGGGGG GGGGSGSGDA QTSSNNINNT SDNKTGDN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 3e-22 | 316 | 392 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the control of processes related to senescence and pathogen defense (PubMed:12000796). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (PubMed:12000796). Activates the transcription of the SIRK gene and represses its own expression and that of the WRKY42 genes (PubMed:12000796). Modulates phosphate homeostasis and Pi translocation by regulating PHO1 expression (PubMed:25733771). {ECO:0000269|PubMed:12000796, ECO:0000269|PubMed:25733771}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00209 | DAP | Transfer from AT1G62300 | Download |
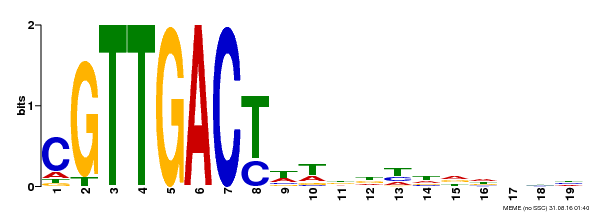
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid, ethylene, jasmonic acid, pathogens, wounding and strongly during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010926185.1 | 0.0 | probable WRKY transcription factor 31 | ||||
| Swissprot | Q9C519 | 1e-141 | WRKY6_ARATH; WRKY transcription factor 6 | ||||
| TrEMBL | A0A2H3YEF3 | 0.0 | A0A2H3YEF3_PHODC; probable WRKY transcription factor 31 | ||||
| STRING | XP_008797553.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5265 | 36 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G62300.1 | 1e-123 | WRKY family protein | ||||



