 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010926296.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 915aa MW: 103582 Da PI: 6.2731 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 182.6 | 4.4e-57 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
l+ ++rwl+++ei++iL+n++k++++ e++++p+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+v+vl+cyYah+een++fqr
XP_010926296.1 20 LEAQRRWLRPTEICEILQNYRKFRIAPEPPNKPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSVDVLHCYYAHGEENENFQR 114
56799****************************************************************************************** PP
CG-1 97 rcywlLeeelekivlvhylevk 118
r+yw+Lee++ +ivlvhylevk
XP_010926296.1 115 RSYWMLEEDYMHIVLVHYLEVK 136
*******************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 85.163 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 9.2E-79 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.8E-50 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.68E-11 | 344 | 415 | IPR014756 | Immunoglobulin E-set |
| PROSITE profile | PS50297 | 19.253 | 497 | 626 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 9.5E-20 | 507 | 627 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 6.37E-19 | 521 | 627 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.83E-16 | 521 | 624 | No hit | No description |
| PROSITE profile | PS50088 | 8.576 | 532 | 564 | IPR002110 | Ankyrin repeat |
| Pfam | PF12796 | 9.0E-8 | 537 | 627 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0042 | 565 | 594 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.897 | 565 | 597 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 560 | 604 | 633 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 8.76E-8 | 737 | 791 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.064 | 740 | 762 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.59 | 741 | 770 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0017 | 742 | 761 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.02 | 763 | 785 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.865 | 764 | 792 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.3E-4 | 766 | 785 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 915 aa Download sequence Send to blast |
MADARRYGLT PQLDIEQILL EAQRRWLRPT EICEILQNYR KFRIAPEPPN KPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHER LKAGSVDVLH CYYAHGEENE NFQRRSYWML 120 EEDYMHIVLV HYLEVKGNKP SFGRTRDVQE TAQVVNMDSP VCSNSFTNHS QLPSQTTDAE 180 SPNSAHTSEY EDAESADNYQ ASSRYNSFLE MQQYGDGPVM DVRLRNPHFP IASINNQCDI 240 QGTQAAEPKS DFNSVAQEDF MRVFDGTGLG LTFSGPRTQY DLTSWDEVLE HCTTGFQTPP 300 FHPGQAAAVE DNPRLETSTG ELYTDELGVK QVDVTTTLDK SLWQILITGT FLKNKEDVEK 360 CQWSCMFGEI EVPAELLADG TLRCHAPLHK SGRVPFYITC SNRLACSEVR EFEFRVDDAQ 420 DMETLDSHGY DTNEMHLHVR LEKLLNLGPV DQQKVVANSV KEHLHLSNKI SSLMMEFDDE 480 WSNLLKLTHE EGFSPDNAKD QLLEKLMKEK LHSWLLHKVA EDGKGPNVLD NEGQGVLHLA 540 AALGYDWAIK PTITSGVNIN FRDVHGWTAL HWAAYCGRER TVVALIALGA APGVLTDPTP 600 EFPTGRTPAD LASANGHKGI AGFLAESSLT NHLSTLTLKE SEGSDIAELS GITDVEDVVE 660 KSAIQVADGD LQAGLSLKDS LTAVRNASLA AARIYQVFRV HSFHRKKLIE YGDDKCGTSD 720 ERALSLIYLK TAKPGQHDMP QHAAAIRIQN KFRGWKGRKE FLIIRQRIVK IQAHVRGHQV 780 RKHHKKIIWT VSIVEKAILR WRRKGSGLRG FRSEGLLEGR STQNQAVKED DYDFLQEGRK 840 QTEARLQKAL ARVKSMVQYP EARDQYRRLV NVVTELQESK AMQDRILNES SEAADGDFMI 900 ELEELWQDDT PMPTA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5hry_A | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| 5hry_B | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| 5hry_C | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| 5hry_D | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| 5hry_E | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| 5hry_F | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| 5hry_G | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| 5hry_H | 2e-11 | 506 | 624 | 43 | 155 | ank3C2_1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
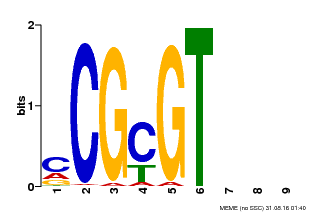
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010926291.1 | 0.0 | calmodulin-binding transcription activator 3 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | A0A3Q0HUC3 | 0.0 | A0A3Q0HUC3_PHODC; calmodulin-binding transcription activator 3-like isoform X1 | ||||
| STRING | XP_008782144.1 | 0.0 | (Phoenix dactylifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.1 | 0.0 | ethylene induced calmodulin binding protein | ||||



