 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010926606.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 806aa MW: 87198.4 Da PI: 7.0692 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 57.9 | 1.9e-18 | 644 | 740 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEE.TTS-EEEEEE...EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS.SEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltled.esgrsWevkliy.rkksgryvltkGWkeFvkangLkegDfvvFkldgr.sef 90
f+kvl++sdv+++gr+vlpkk ae h +++ + ++ + +ed ++++W++++ + ++ks++y+l+ ++ eFv++ngL+egDf++++ ++ +++
XP_010926606.1 644 FQKVLKQSDVGSLGRIVLPKKEAEIHlpELEARDGISIPVEDiGTSQVWNMRF-WpNNKSRMYLLE-NTGEFVRSNGLQEGDFIIIY--SDmKCG 734
99*********************999**999999********7777*******.647777777777.********************..566999 PP
..EEEE CS
B3 91 elvvkv 96
+++++
XP_010926606.1 735 KYMIRG 740
888875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 3.9E-25 | 638 | 749 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.94E-20 | 642 | 736 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10015 | 1.07E-29 | 642 | 741 | No hit | No description |
| Pfam | PF02362 | 6.6E-15 | 644 | 740 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.6E-13 | 644 | 744 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 9.996 | 644 | 744 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 806 aa Download sequence Send to blast |
MEPDGDHLHQ QQHAEAAEEA MAMAAAAATV ATSPAGAADN KDKGKRIWIP DEGPSNPPDD 60 LMLDIEGDDL IFSDAAFSCL PDFPCLPSPA RSPSSSSACN LFHNTSLASS SSSSSSFLFG 120 SDGGGSGGGD LQAVASSSNG ERMMPSEVAS DLLSMELEGL DILGDMGLFD LSEQWNPASL 180 FPDSMTMNGE ATASTSGPGP SPPLHAMAGE VRPPGEAGVG GGAARGAPVG GPEEDPEEMA 240 RVFFEWMRIN RDAISPQDLR EIRMKRATID CAVRHLGGNR QAQVQLLKLV LTWVQANHLQ 300 KKKRRRSQNE HQDRPERAAQ PPYPYHHQHQ QQPFASPNHH LPVMSSDVPV NPNPNLDYSY 360 NADSQRHGWN IPYHVDPTTG VAFPVMMPVY GGGGGSSSGG EPAYPGPAAA ANYSYHQGGT 420 SGIVLNGQPF SPSPDMHAAD HTAAAWPSQY AAAPAQYVPF GGAAGPHPAT MPQAQYPGGF 480 VNQYPGHALY QQGQRLGGMA SVTKEARKKR MARQRRVSSL HSHRSQPQQQ QQQNHHQAES 540 SSSRAPAEGA SSSSSNANNN NNNNNNNNHN NNNNNRTWSF WSSSSSSRQK NPMVETPPPP 600 KSSMQQPQLS PSMPPQQQNS HRDAASSTDR RQGLKGEKNL RFLFQKVLKQ SDVGSLGRIV 660 LPKKEAEIHL PELEARDGIS IPVEDIGTSQ VWNMRFWPNN KSRMYLLENT GEFVRSNGLQ 720 EGDFIIIYSD MKCGKYMIRG IKVRPQPERG RANRSVARSH LKRSSWERGS TSRMRGSSSH 780 DGINDDPGMP SSSAASPFMI KMEASP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9b_A | 6e-35 | 640 | 748 | 1 | 111 | B3 domain-containing transcription factor FUS3 |
| 6j9b_D | 6e-35 | 640 | 748 | 1 | 111 | B3 domain-containing transcription factor FUS3 |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 299 | 305 | QKKKRRR |
| 2 | 300 | 305 | KKKRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
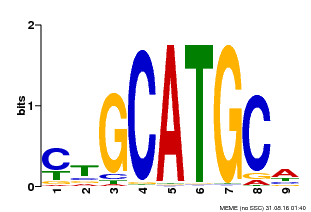
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010926606.1 | 0.0 | regulatory protein viviparous-1 isoform X2 | ||||
| TrEMBL | A0A3Q0HZE8 | 0.0 | A0A3Q0HZE8_PHODC; regulatory protein viviparous-1 | ||||
| STRING | XP_008787720.1 | 0.0 | (Phoenix dactylifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 1e-60 | B3 family protein | ||||



