 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010927134.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 437aa MW: 48543 Da PI: 6.2349 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 103 | 1.9e-32 | 224 | 278 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWt +LHerFveav++L G+ekAtPk +l+lm+v+gLt++hvkSHLQkYRl
XP_010927134.1 224 KPRLRWTLDLHERFVEAVNKLDGAEKATPKGVLKLMNVEGLTIYHVKSHLQKYRL 278
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.067 | 221 | 281 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.8E-31 | 222 | 279 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.2E-17 | 222 | 278 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.9E-24 | 224 | 279 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.0E-8 | 226 | 277 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 2.8E-21 | 315 | 361 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 437 aa Download sequence Send to blast |
MSSHSIITAE QSNSPEGMRH CCHASASSTT NLFNVLSDCQ KLLNDKLSCT SPSSYVQREL 60 INSSSPQKGL LFRLKKSSPE SDPGSPLSNK CEPQNSAVQS LNSPLLHGGN TTNVHNEDEH 120 SDDLMKDFLN LSGDASDSSF HGENYDNNSI ALGEQMELQM LSEQLGIAIT DNGEIPRLDD 180 IYETPAPSVP LSSNCNQTSQ PLRPAAKFQL HSNSSTSTTA TANKPRLRWT LDLHERFVEA 240 VNKLDGAEKA TPKGVLKLMN VEGLTIYHVK SHLQKYRLAK YLPEAKEDKK ASSSEDQKMP 300 TVSHESDPGK KRNIEVIEAL RMQIEVQKQL HEQLEVQRAL QLRIEEHARY LQRILEEQQK 360 ASNTFVFPTQ MPGTEMQLES PHHLSPEQAE SKVDSICSPS SSNSKHKATD SDTRSKPPED 420 HKRARLEVER EGTSLGS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-24 | 224 | 281 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in phosphate starvation signaling (PubMed:26082401). Binds to P1BS, an imperfect palindromic sequence 5'-GNATATNC-3', to promote the expression of inorganic phosphate (Pi) starvation-responsive genes (PubMed:26082401). Functionally redundant with PHR1 and PHR2 in regulating Pi starvation response and Pi homeostasis (PubMed:26082401). {ECO:0000269|PubMed:26082401}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
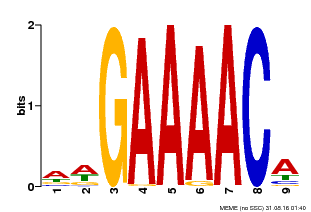
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated under Pi starvation conditions. {ECO:0000269|PubMed:26082401}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_029121639.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 isoform X2 | ||||
| Swissprot | Q6YXZ4 | 1e-100 | PHR3_ORYSJ; Protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| TrEMBL | M0THW1 | 1e-158 | M0THW1_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr7P15740_001 | 1e-158 | (Musa acuminata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 1e-87 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



