 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010930053.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 395aa MW: 45255.8 Da PI: 6.5314 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 85 | 9.4e-27 | 94 | 175 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
W ++e++ Li +rrem+ ++++k++k+lWe++s+km+++gf rsp++C++kw+nl k++kk k+++k+ s+++ +++y+++l+
XP_010930053.1 94 WVQEETRSLISLRREMDGLFNTSKSNKHLWEQISAKMKDKGFDRSPTMCTDKWRNLLKEFKKAKHHAKG--SGAV-KMSYYKELD 175
********************************************************************8..4665.799***998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 8.838 | 86 | 150 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-5 | 87 | 152 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.6E-5 | 90 | 152 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.23E-28 | 92 | 157 | No hit | No description |
| Pfam | PF13837 | 3.8E-21 | 93 | 177 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 395 aa Download sequence Send to blast |
MSLPTLDQYL SQSLLPCFRS REDESDEEQQ GARKSMYLSD KPQSVDYYKE DSRDMMIQVV 60 GNGGLPPPQQ QMILAESSGE DHEVKAPKKR AETWVQEETR SLISLRREMD GLFNTSKSNK 120 HLWEQISAKM KDKGFDRSPT MCTDKWRNLL KEFKKAKHHA KGSGAVKMSY YKELDEMLKE 180 RNKNPAYKSP TVSKVDSYLQ FSDKGSGRST LNMDRRLDHE RHPLAITAAD EVAANGVPPW 240 NWRDTPANGG DNHTSYGGRV ILVKWGDYTR KIGIDGSAEA IKDAIKSAFG IRTRRAFWLE 300 DEDEVVRSLD RDMPLGTYTL HLDDGITIKI CIYDETDRIT VRTEEKTIYT QEDFRDFLNR 360 NGWTGLRELS GFRSVDTMDD LRPGAVYQGI RLLSD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 8e-48 | 88 | 173 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
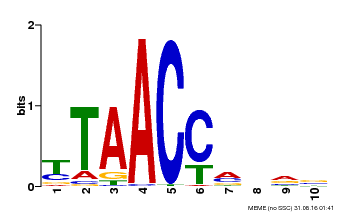
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00640 | PBM | Transfer from MDP0000164819 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010930052.1 | 0.0 | trihelix transcription factor GT-1 | ||||
| Swissprot | Q9FX53 | 1e-174 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A3Q0I2M7 | 0.0 | A0A3Q0I2M7_PHODC; trihelix transcription factor GT-1-like | ||||
| STRING | XP_008792161.1 | 0.0 | (Phoenix dactylifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 1e-171 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



