 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010932273.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1355aa MW: 149749 Da PI: 8.1334 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.9 | 0.00068 | 1239 | 1262 | 1 | 22 |
EEET..TTTEEESSHHHHHHHHHH CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt 22
y C C++sFs+k +L H r
XP_010932273.1 1239 YICDieGCSMSFSTKQDLSLHKRD 1262
889999***************985 PP
| |||||||
| 2 | zf-C2H2 | 12.8 | 0.00036 | 1264 | 1286 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H ++H
XP_010932273.1 1264 CPvkGCGKKFFSHKYLVQHRKVH 1286
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 11.9 | 0.00067 | 1322 | 1348 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
XP_010932273.1 1322 YVCRepGCGQTFRFVSDFSRHKRKtgH 1348
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 2.4E-17 | 35 | 76 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.077 | 36 | 77 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 5.4E-14 | 37 | 70 | IPR003349 | JmjN domain |
| PROSITE profile | PS51184 | 33.22 | 210 | 379 | IPR003347 | JmjC domain |
| SMART | SM00558 | 5.6E-47 | 210 | 379 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 4.39E-25 | 224 | 381 | No hit | No description |
| Pfam | PF02373 | 4.3E-36 | 243 | 362 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 7.8E-4 | 1235 | 1261 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.4 | 1239 | 1261 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.49 | 1262 | 1286 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.967 | 1262 | 1291 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 9.6E-6 | 1263 | 1298 | No hit | No description |
| PROSITE pattern | PS00028 | 0 | 1264 | 1286 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.8E-5 | 1264 | 1290 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| Gene3D | G3DSA:3.30.160.60 | 4.3E-8 | 1291 | 1316 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0029 | 1292 | 1316 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.157 | 1292 | 1321 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1294 | 1316 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 9.41E-10 | 1302 | 1346 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 8.4E-10 | 1317 | 1345 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.845 | 1322 | 1353 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.25 | 1322 | 1348 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1324 | 1348 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1355 aa Download sequence Send to blast |
MAAAAAAGSL GDPTLPPPPP PPPTEVPQWL KSLPLAPEYH PTLAEFQDPI AYILKIEKEA 60 SRFGICKIVP PLPAPPRKTT VANLNRSFAA REPNPKKPPT FTTRQQQIGF CPRRPRPVQK 120 PVWQSGEHYT LQQFETKAKQ FERAYLRKAK KATSSFLSPL EIETLFWKAC ADKPFSVEYA 180 NDMPGSGFAP LGAARRWRGL EEEEEEEAAH VGETAWNMRG VSRAKGSLLR FMKEEIPGVT 240 SPMVYVAMMF SWFAWHVEDH ELHSLNYLHM GAGKTWYGVP RDAALAFEEV VRLHGYGGEV 300 NRLVTFALLG EKTTVMTPEV LMGAGIPCCR LVQNAGEFVV TFPGSYHTGF SLGFNCGEAA 360 NIATPGWLRV AKEAAIRRAS INYPPMVSHF QLLYELALSL CTRMPRGSRS EPRSSRLKDK 420 MKGEGEEIVK NIFVQNVIQN NHLLSFLLDK GSSCIVLPQD APESPLCSNA LVRSQVKAKP 480 RLSLGLCSHQ EAVEASRLLP SDDITPGWSA GRRDMSGLCS LKRNSISVSQ GKMITSATCG 540 RFGAADFYSS SDSQNEEGEK EGTFQGDGLL DQGLLACVTC GILSFACVAV IQPREAAAKY 600 LMSADCGFLS GHINGSGEVS GINKDASRQA DDYNLVSGSG QIGRHTENTV DDYWVHCNTY 660 SGQVSEQSVE VFSDNTGQRG ISALDLLASA YGDTSDTEEE ALHAKSDCTD ENDVKDFPSS 720 AKPIEHPNFA IELPNLCSSK DSQKETDLSF IGADCQNGTC AQNSYNNVGP DDPDGPTNVS 780 AGEKCQLKWE FCGSNQPENV KSAGTDSLDD NRKVTTSNSS IKSMEEPTDF SYREVDDGCR 840 ATETADNQQG NMKMGNASFG SEDLSVQPDV CCDLSEPTKG TAITTQSINV HTKMMNSGIP 900 AMRRCDKDSS RMHVFCLEHA VEVDKQLQPI GGMHIMLFCH PDYPKIEAEA KLLAEELGFD 960 YSWKDVHFKE ATEEEQERFG AALEDEEAIP VNSDWAVKLG INLYYSAGLS KSPLYSKQMP 1020 YNAVIYKAFG CNSPGNSPAK PKTSGRRPGR QKKIVVAGRW CGKVWMTNQV HPYLAHRNEA 1080 QDHDHVEQLY SQGTGQKPKV KTYTKDAPSE RNPSASDGTA ARKSGKKRKK PLRKASKKKP 1140 RHTQIDKSEA MEDVPETSSS PCGRVLRSSH SKQAVIANRK KFNLKDEAEG GPSARLRKQT 1200 SKAMEEVKTK SASRKQARQR KAKKAQATNP VTKDEDGEYI CDIEGCSMSF STKQDLSLHK 1260 RDICPVKGCG KKFFSHKYLV QHRKVHMDDR PLECPWKGCK MTFKWAWART EHIRVHTGDR 1320 PYVCREPGCG QTFRFVSDFS RHKRKTGHSV KKGRR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 2e-71 | 24 | 468 | 3 | 413 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-71 | 24 | 468 | 3 | 413 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 1125 | 1129 | KKRKK |
| 2 | 1125 | 1139 | KKRKKPLRKASKKKP |
| 3 | 1128 | 1138 | KKPLRKASKKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3 with a specific activity for H3K27me3 and H3K27me2. No activity on H3K4me3, H3K9me3, H3K27me1 and H3K36me3. Involved in biotic stress response. May demethylate H3K27me3-marked defense-related genes and increase their basal and induced expression levels during pathogen infection. {ECO:0000269|PubMed:24280387}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
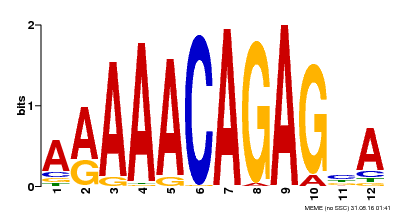
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt stress, abscisic acid (ABA), jasmonic acid (JA), the ethylene precursor ACC and infection by the bacterial pathogen Xanthomonas oryzae pv. oryzae. {ECO:0000269|PubMed:24280387}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010932273.1 | 0.0 | lysine-specific demethylase JMJ705 | ||||
| Swissprot | Q5N712 | 0.0 | JM705_ORYSJ; Lysine-specific demethylase JMJ705 | ||||
| TrEMBL | A0A2H3YKE0 | 0.0 | A0A2H3YKE0_PHODC; lysine-specific demethylase REF6-like isoform X1 | ||||
| STRING | XP_008799926.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5346 | 32 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



