 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010932347.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 386aa MW: 43192.9 Da PI: 4.5885 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.2 | 2.4e-19 | 117 | 166 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+G+r+++ +g+W+AeIrdp++ r++lg+f taeeAa+a++a ++k++g
XP_010932347.1 117 QYRGIRQRP-WGKWAAEIRDPRK---GVRVWLGTFNTAEEAARAYDAEARKIRG 166
69*******.**********954...4*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 7.75E-33 | 117 | 175 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 1.2E-32 | 117 | 175 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.44E-22 | 117 | 176 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 24.131 | 117 | 174 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.3E-38 | 117 | 180 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 7.4E-12 | 118 | 166 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-11 | 118 | 129 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-11 | 140 | 156 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0070483 | Biological Process | detection of hypoxia | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 386 aa Download sequence Send to blast |
MCGGAIISDF IPPGRSWRRV TADCPWPDLK KGVEAKKSRG KRQPPVGIDD DDFETDFQEF 60 NDESPESEVD DEAELIDVKP FAYPSKTPFS RDRSTTLKPA EYDGPVARSA KRKRKNQYRG 120 IRQRPWGKWA AEIRDPRKGV RVWLGTFNTA EEAARAYDAE ARKIRGKKAK VNFPDEMRPS 180 AQKRSQKAAA PRATESNQPE ILKFNQSLSY LDEPHQDLYS TFDFIEEKQP IKQAGNLSAF 240 PAINPALPTE GPGTNLYSDQ GSNSFGCSDL GWEHEAKTPE ITSVLALIPT IIEGEQSAFL 300 EDGAPQKKMK NNAGEAVPPS EENAAVKLSE DMSDFDSFLK FLQIPYPEES SDESIDHLLS 360 SDVTQDMNDV DLWSFDYLPA VEGSVY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 9e-21 | 116 | 176 | 13 | 74 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00204 | DAP | Transfer from AT1G53910 | Download |
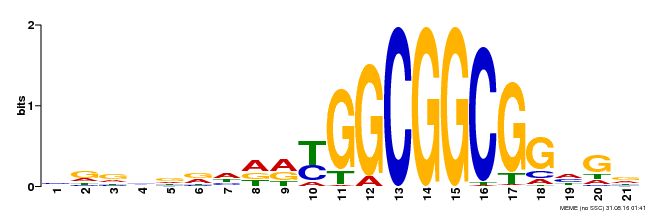
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010932347.1 | 0.0 | ethylene-responsive transcription factor 1 | ||||
| TrEMBL | A0A2H3XIM2 | 0.0 | A0A2H3XIM2_PHODC; ethylene-responsive transcription factor 1-like | ||||
| STRING | XP_008784363.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1039 | 38 | 134 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G14230.3 | 9e-61 | related to AP2 2 | ||||



