 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010938302.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 897aa MW: 100520 Da PI: 6.6931 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 126 | 1.5e-39 | 3 | 84 | 36 | 117 |
CG-1 36 sgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylev 117
g+++L++r ++r+frkDG++wkkkkdgktv+E+hekLK+g+ e +++yYa+s+++p+f rrcywlL+++le+ivlvhy+++
XP_010938302.1 3 GGTILLFDRTMLRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNEERIHVYYARSDDDPNFYRRCYWLLDKNLERIVLVHYRQT 84
699****************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01076 | 1.8E-33 | 1 | 85 | IPR005559 | CG-1 DNA-binding domain |
| PROSITE profile | PS51437 | 57.031 | 1 | 90 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 6.8E-34 | 3 | 83 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.01E-12 | 341 | 427 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 9.93E-14 | 512 | 642 | No hit | No description |
| Pfam | PF12796 | 2.1E-6 | 530 | 611 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-14 | 532 | 648 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.64E-14 | 533 | 648 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.239 | 550 | 615 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.568 | 583 | 615 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1.6E-5 | 583 | 612 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 4100 | 622 | 651 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.49E-7 | 698 | 802 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.144 | 733 | 762 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 3 | 751 | 773 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.675 | 752 | 781 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0014 | 754 | 772 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 1.6E-4 | 774 | 796 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.182 | 775 | 799 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.0E-5 | 776 | 796 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 1.1 | 855 | 877 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.157 | 857 | 885 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.092 | 858 | 877 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 897 aa Download sequence Send to blast |
MNGGTILLFD RTMLRNFRKD GHNWKKKKDG KTVQEAHEKL KIGNEERIHV YYARSDDDPN 60 FYRRCYWLLD KNLERIVLVH YRQTSENPPT LVECTDASSL TNRMHHGSPS TPMISSGSAR 120 SELSGSAVMS EEINSREDRA INTGSGISLS DKYNELRNQE FSLHDINTLD WEDLVEPPTS 180 TVSVLCREGD ALSSVQQMPD GLRNSVNNLS VLPSHDVVEG VPSSGHPTDI TNDNGIDGVN 240 CSGYFQAAKD QENPSLLFET MNSTIQEADF KLDSAHSSAS PDIFTGDVFL THNSFGGWNS 300 MNEDSFGLVT EQLEALNSSG DKSNAFTIMD QSSTTEQIFS ISDISPGWAY STEETKVLVV 360 GHFTDPYKHP MTSNIYCAFG EMHAAAELIQ AGVYRCTAMP HPPGSVNFFM TLDGYTPISQ 420 VLSFDYRSVP SDKLDGGVTS SEDDNNNLKG KEFQVQTRLA HLLFSTTNNI SIQSSRTQSK 480 SLKEAKRYAS VTSPLMEKDW MNLLRLSSNG EAFHVPATKD LFELVLKNKL QEWLLEKVAE 540 GCKTPAHDSQ GQGVIHLCAI LDYAWAAHLF SLSGMSLDFR DASGWTALHW AAYFGREKMV 600 AALLSAGANP SLVTDPTPEF PGGCIAADLA SNQGFEGLAA YLAEKGLTAH FQAMSLSGNI 660 SAPLLPSSTD KSSSENLYPE NLTEQELCLR ESLAAYRNAA DAADRIQAAF RERALKQQTK 720 AVQLVKPEIE AAQIVAALKI QHAFRNYSRR RIMKAAARIQ SNFRTWKTRR HFLNMRKHAI 780 KIQAAFRGHQ VRKQYHKIIW SVGVLEKAIL RWRLKRKGLR GVQVESKEAM KVDSNEESTG 840 EEEFFRISRE QAEERVKRSV IRVQAMFRSH RAQQEYRRMK LAYEQANLEF NELDQRP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
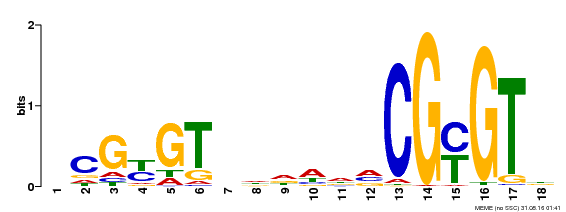
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010938301.1 | 0.0 | calmodulin-binding transcription activator CBT isoform X2 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A2H3XN33 | 0.0 | A0A2H3XN33_PHODC; calmodulin-binding transcription activator CBT isoform X2 | ||||
| STRING | XP_008786234.1 | 0.0 | (Phoenix dactylifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||



