 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010938858.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 860aa MW: 96043.7 Da PI: 5.6966 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 114.7 | 5e-36 | 166 | 242 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cqv++C ad+ e+k yhrrh+vC +++a++v+++g+ +r+CqqC++fh ls+fDe+krsCrr+L++hn+rrr+k
XP_010938858.1 166 TCQVPACGADIRELKGYHRRHRVCLRCANASSVVLDGEPKRYCQQCGKFHLLSDFDEGKRSCRRKLERHNRRRRRKP 242
6************************************************************************9975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.6E-28 | 162 | 228 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.282 | 164 | 241 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.44E-32 | 165 | 244 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.4E-26 | 167 | 241 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 860 aa Download sequence Send to blast |
METDLNPVTA VPSPAPSLTP SSPRVRVSEM EIPPMADGSA AAAEAEAAVW DWENLLDCTI 60 DEEDPWNLFW GSTEHGQTST PPPLPPLMAG EPSNLPPVPA GTAAADGNNR VRKRDLRLVC 120 PNYLAGRVPC ACPEMDEMEL EEEDVGEVVA GGRKRARTGG VAAAATCQVP ACGADIRELK 180 GYHRRHRVCL RCANASSVVL DGEPKRYCQQ CGKFHLLSDF DEGKRSCRRK LERHNRRRRR 240 KPSDSRNAVE KELETQGDPQ TDVICNGKPG KDMSNIACDT VETVVCDRSL DAETLLGSED 300 GSGSPICSIP SFQNDQNASL ASFAASAEAH ADERRDNSKS ALSSTLCDNK STYSSVCPTG 360 RISFKLYDWN PAEFPRRLRH QIFHWLASMP VELEGYIRPG CTILTVFISM PQFMWEKLSQ 420 DVAQCIRDLV NAPGSLLLGR GTFFIYLSHM IVHVLKDGTS LMNIKMEVQA PRLHYVYPTY 480 FEAGKPMEFV ACGSNLNQPK FRFLVSFAGK YLSFDSCSVI SHGKTKYYDE NGAECLHNSD 540 HEMLRINITE TNLDVFGPAF IEVENVYGIS NFVPILVGNK NVCSEMKRIQ DALGEPYCAD 600 SIMSQNAVSG ASPALCEFFV SRQNAISELL LDIAWLLKEP HIDERESLFS LGDIQRLTSL 660 LKFLIQNRFV SIIEVVLHYW DNIVDGKGFD KSENWYSDAD MRLLHEYVNH AKEILCQRTL 720 HDVESELDSK NSIDGNDVLQ SYLQSDVVYT LPCTNQDRGA RNEDSSCSAT SSATQEDDEN 780 VALVTNRIIH RRRCYPHLGT QWHRQSWGDI FPTNIMRRRL GVILMVSVVT CFAACIVLFH 840 PHRAREFAVF RRCLFGGPTQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 2e-39 | 162 | 245 | 1 | 84 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 223 | 241 | KRSCRRKLERHNRRRRRKP |
| 2 | 227 | 239 | RRKLERHNRRRRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
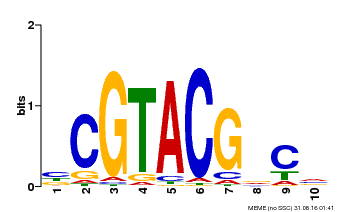
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010938858.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A2H3Z6X2 | 0.0 | A0A2H3Z6X2_PHODC; squamosa promoter-binding-like protein 9 isoform X1 | ||||
| STRING | XP_008809291.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.1 | 1e-174 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



