 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010940145.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 726aa MW: 83160.8 Da PI: 6.5736 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 76.6 | 5.1e-24 | 103 | 187 | 2 | 90 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHela 90
YneYAk++GF vrk+ ++sk+n+e++ +++vC++egk + ++k+ + +++ rtgC a lkvk++ +++w+v ++ +eHnHel
XP_010940145.1 103 LYNEYAKDMGFGVRKKLCRRSKTNNEMIAAWYVCTREGKIAASEKA----ECSQSARRTGCCAALKVKRMANKEWKVVHFAKEHNHELD 187
7****************************************99888....889999*******************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 9.0E-21 | 103 | 188 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 3.9E-32 | 299 | 392 | IPR018289 | MULE transposase domain |
| Pfam | PF04434 | 2.0E-9 | 590 | 625 | IPR007527 | Zinc finger, SWIM-type |
| PROSITE profile | PS50966 | 10.423 | 590 | 626 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 9.1E-8 | 601 | 628 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 726 aa Download sequence Send to blast |
MESTNDGSSQ KLANVDDIEL AAETGLADDD DDDDDDDDDD EEEEEGEEAS IIRDSAAIEL 60 GDPNTSTDLE VNSLAGNLEE VGGTRSEPSI GMMFQSAVAA YRLYNEYAKD MGFGVRKKLC 120 RRSKTNNEMI AAWYVCTREG KIAASEKAEC SQSARRTGCC AALKVKRMAN KEWKVVHFAK 180 EHNHELDPEN VCLFRSHMKN RMHNKNRINK AGVGHKYTTS ASSKQSGGQD GECQNLVDRG 240 QRRFFAPADA HAIHMLFIHM HSKNPAFFYA VDFDEEQHLK NVLWADAKAK VAYTHFGDVV 300 TFDTTYLTDG YKLPFAPFVG VNHHGQTVLL GCALLADQST ASFVWLFKTW LDAMSGRPPK 360 AIITDYNKDI AAAMSQLFPE THHRYCLWHI LKRVPEKLGD VCEARENFLK KLNKCIYDST 420 TAVEFERRWW NLIHRFGLAN EEWLQSLYKD RQQWVPLYLK DTFFAGMSIS QRNESVSTIF 480 HKFITRETGL KEFLDKYEVA LESKFEDEAQ EDFWSFHSKT KEITHPSPFE LQLADVYTRN 540 MFEKFQMEVL GISSVFASKV EENGDIATYM VKAYEVQEKM KARRLLEKEY QVVWNAIERK 600 IRCACRLFEF KGYLCRHALA VFLALGVLEV PSSYILKRWT KDAKSRHVLD EGYTVHGDCS 660 ESVAQRFNDI CVRSFKFAEE ASLTKRSYDV ALLALQEAFR RVTHANNSVR KSRQRNYPHN 720 GGRSNK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
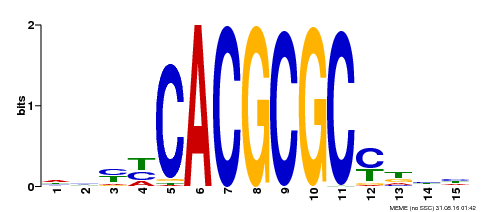
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010940145.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A3Q0IA20 | 0.0 | A0A3Q0IA20_PHODC; protein FAR-RED IMPAIRED RESPONSE 1-like isoform X1 | ||||
| STRING | XP_008796002.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP18 | 32 | 967 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 1e-173 | FAR1 family protein | ||||



