 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010940930.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 342aa MW: 37942.1 Da PI: 9.857 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 367.7 | 2.1e-112 | 1 | 342 | 1 | 301 |
GAGA_bind 1 mdddgsre..rnkg.yye........paaslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllal 84
mdd +re r k+ +++ p++++k++++++lm+++aerd++i++rnla+sekkaa+aerdma lqrd+a+ ern+a++erdn+++al
XP_010940930.1 1 MDDGRHREngRPKPdQFKsvhaqwmmPQHQMKDHQTMKLMAIMAERDSAIHDRNLAISEKKAALAERDMAILQRDAAIVERNNAFLERDNAIAAL 95
78887777788888888899999988778****************************************************************** PP
GAGA_bind 85 llvensla.....salpvgvqvlsgtksidslqqlse..pqledsave.lreeeklealpieeaaeeakekkkkkkrqrakkp..kekkakkkkk 169
++++++ + + + + ++ tk+ d+l +++ +ql+d +++ r+ ++ ea+pi++a+e+ak k k+ r+++ ++ kk+k++kk
XP_010940930.1 96 QYARETGMngnggN--GCSPGCTAPTKHHDHLPHVHPppSQLSDAPYDhERQVDIAEAYPISTAVESAKGCKAKRMRKENGVQaiPIKKSKSPKK 188
*9877766354333..333345566777777777666889********99999***********9999876666555544433113455556666 PP
GAGA_bind 170 ksekskkkvkkesader......................skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvy 242
++++ +++ +k+ + + +k+++k +dl+ln+vs+Dest+PvPvCsCtG+ + CYkWGnGGWqS+CCttt+S+y
XP_010940930.1 189 NKRSGNDDLNKQVSIAKfhgewkgqeqvsgggdankipvTKHHWKGQDLGLNQVSFDESTMPVPVCSCTGKFHPCYKWGNGGWQSSCCTTTLSMY 283
5555555555555444478899************************************************************************* PP
GAGA_bind 243 PLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
PLPv++++r+aR++grKmS+gaf klL++LaaeG+dls+pvDLkdhWAkHGtn+++ti+
XP_010940930.1 284 PLPVMPNKRHARVGGRKMSGGAFIKLLSRLAAEGHDLSTPVDLKDHWAKHGTNRYITIK 342
**********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 1.3E-145 | 1 | 342 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 1.1E-98 | 1 | 342 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 342 aa Download sequence Send to blast |
MDDGRHRENG RPKPDQFKSV HAQWMMPQHQ MKDHQTMKLM AIMAERDSAI HDRNLAISEK 60 KAALAERDMA ILQRDAAIVE RNNAFLERDN AIAALQYARE TGMNGNGGNG CSPGCTAPTK 120 HHDHLPHVHP PPSQLSDAPY DHERQVDIAE AYPISTAVES AKGCKAKRMR KENGVQAIPI 180 KKSKSPKKNK RSGNDDLNKQ VSIAKFHGEW KGQEQVSGGG DANKIPVTKH HWKGQDLGLN 240 QVSFDESTMP VPVCSCTGKF HPCYKWGNGG WQSSCCTTTL SMYPLPVMPN KRHARVGGRK 300 MSGGAFIKLL SRLAAEGHDL STPVDLKDHW AKHGTNRYIT IK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
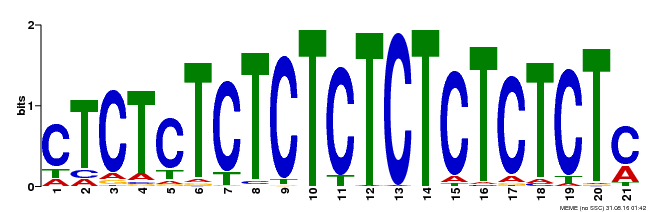
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010940930.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_010940931.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_010940932.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_010940933.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_010940935.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_010940936.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_019711083.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_029116129.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_029116130.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_029116131.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_029116132.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_029116133.1 | 0.0 | barley B recombinant-like protein D | ||||
| Swissprot | Q5VSA8 | 1e-139 | BBRD_ORYSJ; Barley B recombinant-like protein D | ||||
| TrEMBL | A0A2H3ZHQ5 | 0.0 | A0A2H3ZHQ5_PHODC; barley B recombinant-like protein D isoform X1 | ||||
| STRING | XP_008776328.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6226 | 37 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 1e-110 | basic pentacysteine 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



