 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010941148.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 628aa MW: 69162.1 Da PI: 6.6708 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 109.2 | 2.7e-34 | 432 | 516 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW++ ev aLi++r+++e+r+++ lk+plWeevs+ m++ g++rs+k+Ckekwen+nk+++k+k+++kkr +++s+tcpyf+ql+
XP_010941148.1 432 RWPRAEVEALIQVRSRLESRFQEPGLKGPLWEEVSATMAAMGYHRSAKRCKEKWENINKYFRKTKDSSKKR-PRNSKTCPYFHQLD 516
8*********************************************************************8.9999*********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 7.259 | 425 | 489 | IPR017877 | Myb-like domain |
| CDD | cd12203 | 2.79E-28 | 431 | 496 | No hit | No description |
| Pfam | PF13837 | 1.1E-24 | 431 | 518 | No hit | No description |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 628 aa Download sequence Send to blast |
MQSGYGSVSE MQQFMMESCG SSIFSISAAS PNPSADIHAA APQPLKYHPL HPHHHHQPPH 60 PQPPPPPPHH FSPFHPIPIT QQLFQQSHQF QLLQHQHENS AGATAAAGGG PSFLAPAMNF 120 KLAANESSGG GSHEGLNDDD GGSESRLHHH WQREEESAIK EPSWRPLDID YINRNNKRCK 180 EKETETPTNK YSKKTKEGAE PDHGGHITGG SNYKLFSELE AICKPGVCSL GGGGGANQTG 240 SGSALTGDET ALMPTATNPP GILTADHHVG GSETSAGEEA TARKFSKGSG RKKRKRRQQK 300 KQQLSSVMAF FENLVKQLME HQENLHQKFL EVMERRDQER TLREEAWRRQ EAAKSSHEAA 360 ARAHDRVLAS SREAAIISFL EKTTGETLHL PEKLRFPSQL SEEPGKLEAE TTQNPPTEPS 420 NNANKVPFNT SRWPRAEVEA LIQVRSRLES RFQEPGLKGP LWEEVSATMA AMGYHRSAKR 480 CKEKWENINK YFRKTKDSSK KRPRNSKTCP YFHQLDQLYS KSLNKSHPAS SSSPNANVAA 540 GGAIASGTAG DRRKDNSELL DAIVVSTDQQ GFKFPEMSSL HDFGFNGKGD DNSELHRNVG 600 SNGEEDDEED EEEGGGGEEG EEREEGEG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 282 | 297 | RKFSKGSGRKKRKRRQ |
| 2 | 289 | 297 | GRKKRKRRQ |
| 3 | 290 | 295 | RKKRKR |
| 4 | 290 | 296 | RKKRKRR |
| 5 | 290 | 297 | RKKRKRRQ |
| 6 | 291 | 296 | KKRKRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00651 | PBM | Transfer from LOC_Os02g01380 | Download |
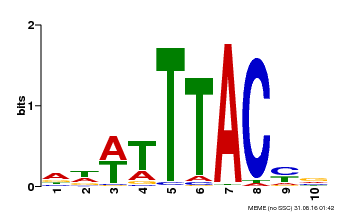
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010941148.1 | 0.0 | trihelix transcription factor GT-2 | ||||
| TrEMBL | A0A2H3Y811 | 0.0 | A0A2H3Y811_PHODC; trihelix transcription factor GTL1-like | ||||
| STRING | XP_008794787.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6399 | 35 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 3e-55 | Trihelix family protein | ||||



