 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010943421.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 296aa MW: 33455.1 Da PI: 7.9462 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 91.9 | 5.5e-29 | 186 | 235 | 2 | 51 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQ 51
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ
XP_010943421.1 186 PRMRWTTTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQ 235
9************************************************* PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.22E-13 | 183 | 236 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-25 | 184 | 235 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.0E-20 | 186 | 235 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.0E-6 | 187 | 235 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 296 aa Download sequence Send to blast |
MELFPAQPDL SLQISPPNSN PTPSWRKANE NLDLGFWRRA MDSNTTTTTN PFTTTSNLHS 60 ITNRSIAEPD NTAFELSLAH PSAIDSTTMS NSTLLHHPHH HHHHLHLHHH HHHPLVHEGY 120 HQDLGLMRPI RGIPVYQHQP NPFPLVPQLQ QQSLCDSSTA TNFFPFASRS RFLSSRFPVK 180 RSMRAPRMRW TTTLHARFVH AVELLGGHER ATPKSVLELM DVKDLTLAHV KSHLQANLMG 240 LRMDQLEKFL MTICLISTST VEQNRQLSME GQLCTMETIT VVYGAILQAG EAGIMA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 4e-14 | 187 | 235 | 4 | 52 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that regulates lateral organ polarity. Promotes lateral organ abaxial identity by repressing the adaxial regulator ASYMMETRIC LEAVES2 (AS2) in abaxial cells. Required for abaxial identity in both leaves and carpels. Functions with KAN2 in the specification of polarity of the ovule outer integument. Regulates cambium activity by repressing the auxin efflux carrier PIN1. Plays a role in lateral root formation and development. {ECO:0000269|PubMed:11395775, ECO:0000269|PubMed:11525739, ECO:0000269|PubMed:14561401, ECO:0000269|PubMed:15286295, ECO:0000269|PubMed:16623911, ECO:0000269|PubMed:17307928, ECO:0000269|PubMed:18849474, ECO:0000269|PubMed:20179097}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
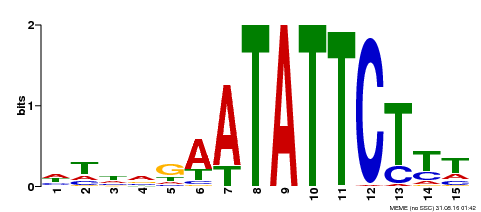
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by AS2 in adaxial tissue. {ECO:0000269|PubMed:18849474}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010943421.1 | 0.0 | probable transcription factor KAN2 isoform X2 | ||||
| Swissprot | Q93WJ9 | 3e-34 | KAN1_ARATH; Transcription repressor KAN1 | ||||
| TrEMBL | A0A3Q0IEJ5 | 1e-119 | A0A3Q0IEJ5_PHODC; probable transcription factor KAN2 | ||||
| STRING | XP_008802564.1 | 1e-120 | (Phoenix dactylifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17695.1 | 3e-34 | G2-like family protein | ||||



