 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010943442.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Arecoideae; Cocoseae; Elaeidinae; Elaeis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 415aa MW: 43878.8 Da PI: 6.296 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 70.8 | 2e-22 | 302 | 364 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr+rrkq+NRe+ArrsR+RK+ae+eeL++++++L++eN++L+ e+++++ke+++l s++
XP_010943442.1 302 ERELKRQRRKQSNRESARRSRLRKQAECEELAQRADSLKEENSSLRAEVNRIRKEYEQLLSQN 364
89**********************************************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 1.5E-32 | 1 | 98 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 4.2E-50 | 136 | 274 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 9.6E-20 | 295 | 363 | No hit | No description |
| Pfam | PF00170 | 1.3E-20 | 302 | 364 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 1.0E-22 | 302 | 366 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.679 | 304 | 367 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.75E-11 | 305 | 362 | No hit | No description |
| CDD | cd14702 | 7.53E-25 | 307 | 357 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 309 | 324 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MGNSEADTSA KTPKTSASQD GSPASSASPA ATVFPDWTSF QAYSPIPPHG FFHSSVASSP 60 QAHPYMWGAQ QFMPPYGTPP HPYVMYPPGG LYAHPSMPPG SHPFNPYAMP SPNGNAETGA 120 VPGGTEMDGK SSEGKERSPL KRSKGSLGSL NMITGKNNNE PGKTSGASAN GGFSQSGESG 180 TESSSEGSDA NSQNDSQPKT SGGQESFDET SHRGNHTPGS QNGGTRMLSQ AVLGQTIPIM 240 PMPAAGATSG VAGPTTNLNI GMDYWGAPSS SSLPPMHGKV PASAVAGAVV PGAPSELWMQ 300 DERELKRQRR KQSNRESARR SRLRKQAECE ELAQRADSLK EENSSLRAEV NRIRKEYEQL 360 LSQNTSLKER LGEMQQGTDN PRLDGDEQNS GDDNHKRHSD SDAQAGEKDP VQSGL |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 318 | 324 | RRSRLRK |
| 2 | 318 | 325 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in responses to fungal pathogen infection and abiotic stresses. {ECO:0000269|Ref.1}. | |||||
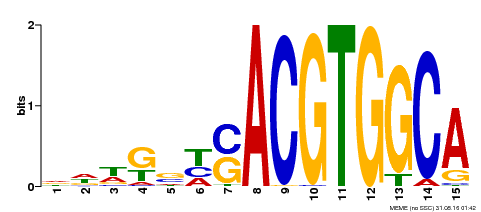
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00291 | DAP | Transfer from AT2G35530 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced during incompatible interaction with the fungal pathogen Puccinia striiformis (Ref.1). Induced by abscisic acid (ABA), ethylene, cold stress, salt stress and wounding (Ref.1). {ECO:0000269|Ref.1}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010943443.1 | 0.0 | bZIP transcription factor 16 isoform X1 | ||||
| Refseq | XP_010943444.1 | 0.0 | bZIP transcription factor 16 isoform X1 | ||||
| Swissprot | B6E107 | 1e-156 | BZP1B_WHEAT; bZIP transcription factor 1-B | ||||
| TrEMBL | A0A2H3ZZL2 | 0.0 | A0A2H3ZZL2_PHODC; bZIP transcription factor 16-like isoform X1 | ||||
| STRING | XP_008810162.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2147 | 37 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G35530.1 | 1e-129 | basic region/leucine zipper transcription factor 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



