 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011070054.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 296aa MW: 33952.1 Da PI: 9.6422 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 18.1 | 7.4e-06 | 26 | 50 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp +C+ s++rk++L+rH+ H
XP_011070054.1 26 FTCPvdNCHSSYRRKDHLTRHMIQH 50
89********************887 PP
| |||||||
| 2 | zf-C2H2 | 18.1 | 7.5e-06 | 55 | 80 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp C+++F + n+krHi+ H
XP_011070054.1 55 FECPieECKRRFAFQGNMKRHIKElH 80
89*********************988 PP
| |||||||
| 3 | zf-C2H2 | 20.2 | 1.5e-06 | 94 | 118 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y+Cp Cgk+F+ s L +H +H
XP_011070054.1 94 YVCPelGCGKVFKYLSKLSKHEDSH 118
9********************9888 PP
| |||||||
| 4 | zf-C2H2 | 14.3 | 0.00012 | 185 | 209 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C +F + snL++H++ H
XP_011070054.1 185 KCSfkGCLHTFATESNLNQHMKAvH 209
688889***************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 1.49E-6 | 23 | 52 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-8 | 23 | 52 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.598 | 26 | 55 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0048 | 26 | 50 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 28 | 50 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 7.21E-6 | 53 | 81 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 7.6E-6 | 53 | 77 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.023 | 55 | 80 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.658 | 55 | 85 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 57 | 80 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 7.6E-7 | 90 | 118 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0028 | 94 | 118 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.72 | 94 | 123 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 96 | 118 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.3 | 126 | 151 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 128 | 151 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.5E-4 | 128 | 149 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 23 | 154 | 175 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 1.68E-7 | 167 | 222 | No hit | No description |
| PROSITE profile | PS50157 | 12.279 | 184 | 214 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0011 | 184 | 209 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 9.6E-6 | 185 | 211 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 186 | 209 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-4 | 212 | 237 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.4 | 215 | 241 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 217 | 241 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 296 aa Download sequence Send to blast |
MQEASSTLNG LVPGGRPPEK LCQRPFTCPV DNCHSSYRRK DHLTRHMIQH QGKLFECPIE 60 ECKRRFAFQG NMKRHIKELH QESSSTNVEP PKEYVCPELG CGKVFKYLSK LSKHEDSHVK 120 LDTAGAFCSE PGCMKYFTNK QSLKEHLCSC HQYIVCDKCG SKQLKKNFKR HLSMHEAAIS 180 MERIKCSFKG CLHTFATESN LNQHMKAVHL GLKPYACSIP GCHMKFAFKH VRDKHKKSGY 240 HVYTPGNFEE SDEQFRSRPR GGRKRKRPTI ETLLRKRVVP PTNSDVSKLL SADSDD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 257 | 265 | RPRGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
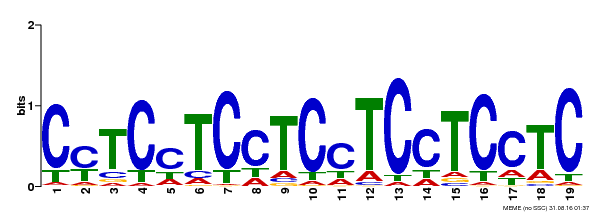
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011070054.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011070054.1 | 0.0 | transcription factor IIIA isoform X2 | ||||
| Swissprot | Q84MZ4 | 7e-82 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A068V6C1 | 1e-139 | A0A068V6C1_COFCA; Uncharacterized protein | ||||
| STRING | VIT_08s0040g00240.t01 | 1e-137 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.1 | 9e-99 | transcription factor IIIA | ||||



