 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011071308.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 401aa MW: 45137.9 Da PI: 6.3638 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 83.5 | 2.7e-26 | 79 | 163 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
W ++e+++Li +r+e++ ++++k++k+lW+ +s kmre+gf rsp++C++kw+nl k++kk k+++++++ + s +++y++++e
XP_011071308.1 79 WVQEETRTLISLRKEIDMLFNTSKSNKHLWDNISLKMREKGFDRSPTMCTDKWRNLLKEFKKAKQNNQDGNGNGSAKMSYYKEIE 163
***********************************************************************9***********98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.3E-5 | 70 | 140 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 8.49 | 71 | 135 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 3.2E-5 | 75 | 137 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 4.41E-26 | 77 | 142 | No hit | No description |
| Pfam | PF13837 | 1.5E-19 | 78 | 165 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 401 aa Download sequence Send to blast |
MYMSEKPQQQ QESAIDFYNS GKQEPPHQNN NMIIQLPSQP HHHLADGGAT TPSGGITATN 60 HNSSNPSSSS APKKRAETWV QEETRTLISL RKEIDMLFNT SKSNKHLWDN ISLKMREKGF 120 DRSPTMCTDK WRNLLKEFKK AKQNNQDGNG NGSAKMSYYK EIEEILRERS KNGPSWKNSE 180 ASNNAGGAGA SKVDSFMQFA DKGIDDTGLT FGPVEANGRS ALNLERRLDH DGHPLAITAA 240 DAVGASGVSP WNWRETPGSG EQTNTYDGRV ITVKLGDYTK RIGIDGSAEA IKEAIKSAFR 300 LRTRRAFWLE DEDNVVRSLD RDMPLGNYTL HVDEGLTIKV CFYEESDPLP VHTEDKTFYT 360 EDDFREFLSR RGWTCLREYN GYRNFDSMDE LCPGAIYRGS N |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 7e-42 | 73 | 161 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00640 | PBM | Transfer from MDP0000164819 | Download |
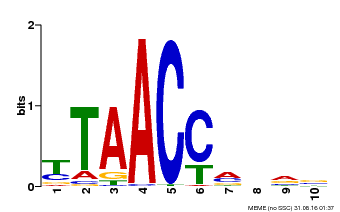
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011071308.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011071308.1 | 0.0 | trihelix transcription factor GT-4-like | ||||
| Swissprot | Q9FX53 | 1e-169 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A2G9I9G9 | 0.0 | A0A2G9I9G9_9LAMI; Transcription factor GT-2 | ||||
| STRING | Migut.E00236.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5113 | 21 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G25990.1 | 1e-168 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



