 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011082155.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 686aa MW: 77711.2 Da PI: 6.7274 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 138.7 | 5.7e-43 | 66 | 222 | 3 | 142 |
DUF822 3 sgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyr...............kgskpl.eeaea 81
+ rk+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+V++AL+reAGw+ve+DGttyr ++ +++ +a
XP_011082155.1 66 KSRKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVIAALAREAGWTVEADGTTYRhsspapaaqapmaqfNSAQQNnINAPN 160
789*****************************************************************544444333333322122222233333 PP
DUF822 82 agssasaspesslq.sslkssalaspvesysaspksssfpspssldsislasaasllpvlsv 142
+g+ +s+es+l ss+k+++++s v+ + + ++ ++ +sp+slds+ ++ +a+l +++s
XP_011082155.1 161 MGAHPVRSVESPLYsSSVKNCSMRSSVDCQPSALRVDENLSPASLDSVAAERDAKLDKYSSA 222
44444466677766577788888888888888888888888888888877766666655554 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 7.3E-46 | 66 | 226 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 2.69E-158 | 250 | 684 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.1E-169 | 252 | 683 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.6E-79 | 274 | 645 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 289 | 303 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 310 | 328 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 332 | 353 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 425 | 447 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 498 | 517 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 532 | 548 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 549 | 560 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 567 | 590 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.1E-53 | 605 | 627 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 686 aa Download sequence Send to blast |
MNTNVNTMNH SFAAGAQDLE PQQSPPPPTT TSHPYHPQPQ PQRRPRGFAA TNSNNNTTIS 60 SSTPTKSRKE REKEKERTKL RERHRRAITS RMLAGLRQYG NFPLPARADM NDVIAALARE 120 AGWTVEADGT TYRHSSPAPA AQAPMAQFNS AQQNNINAPN MGAHPVRSVE SPLYSSSVKN 180 CSMRSSVDCQ PSALRVDENL SPASLDSVAA ERDAKLDKYS SATAINSPEC LEARQLVQDV 240 HCREHGNDLS GTSYIPVYVR LATGLINNFC QLMDPEGIKE ELQHLKALHV DGIVVDCWWG 300 IVEAWAPKKY EWAGYREMFN IIREFRMKLQ VIMAFHEYGG HDSGGIFISL PQWVLEIGKS 360 NQDIFFTDRE GRRNTECLSW GIDKERVLRG RTGIEVYFDF MRSFRTEFND LFTEGLISAV 420 EIGLGASGEL KYPAFPERMG WRYPGIGEFQ CYDKYLLQNL EKAAKLRGHS FWGRGPDNAG 480 YYNSKPHETG FFCERGDFDS YYGRFFLWWY TKVLIDHADN VLSLASLAFE DIQIVVNIPA 540 VYWWYKTSSH AAELTAGYYN SSNRDGYSPL FEVLKKHAAR IKFVISGMQA SYQEIDDALS 600 DPEALSWQVL NAAWDQGLSV AGQNGQPCYD REGFTRLVET ARPRNNPDCR HFSFFVFQQP 660 SPLIQRTICF SEIEYFIKSM HGDVSN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1v3i_A | 1e-131 | 253 | 683 | 10 | 444 | Beta-amylase |
| 1wdq_A | 1e-131 | 253 | 683 | 10 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
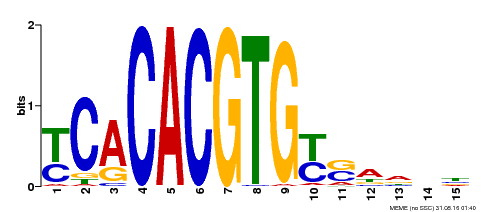
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011082155.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011082155.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A4D9BKE8 | 0.0 | A0A4D9BKE8_SALSN; Uncharacterized protein | ||||
| STRING | Migut.A00486.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1607 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



