 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011085824.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 259aa MW: 29940.3 Da PI: 4.5528 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.8 | 5.7e-21 | 64 | 117 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t++q++ Le+ Fe+++++ e+++eLAkklgL+ rqV +WFqNrRa++k
XP_011085824.1 64 KKRRLTPDQVNLLEKSFEEENKLEPERKTELAKKLGLQPRQVAIWFQNRRARWK 117
5678*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 134 | 5.4e-43 | 63 | 153 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ +qv+lLE+sFeee+kLeperK+ela++Lglqprqva+WFqnrRAR+ktkqlE+dy++Lk++yd+l+++ +++ ke+++L++el
XP_011085824.1 63 EKKRRLTPDQVNLLEKSFEEENKLEPERKTELAKKLGLQPRQVAIWFQNRRARWKTKQLERDYDHLKSSYDSLVSNYDSIIKENNKLKAEL 153
69**************************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.71E-20 | 56 | 121 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.204 | 59 | 119 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.3E-20 | 62 | 123 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 3.5E-18 | 64 | 117 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.86E-18 | 64 | 120 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-22 | 65 | 126 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.1E-6 | 90 | 99 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 94 | 117 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.1E-6 | 99 | 115 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 6.2E-16 | 119 | 161 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 259 aa Download sequence Send to blast |
MDSGRIFFDS SSNGNLLFLG NGDSIFRGGS RVMNMGESSK RRPFFGTAED LYDEEYYDEQ 60 LPEKKRRLTP DQVNLLEKSF EEENKLEPER KTELAKKLGL QPRQVAIWFQ NRRARWKTKQ 120 LERDYDHLKS SYDSLVSNYD SIIKENNKLK AELLFLTEKL QPKESTQEEG FDIQNPNPTP 180 APYLQLIMKV EDRLSTGRQR RGQLLDSGDS YLEYQQGCMA SVEGVKSEED EGSEEGGSYF 240 SDVFAAPEPQ EPVGWFMWP |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 111 | 119 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription activator involved in leaf development. Binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:8535134}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
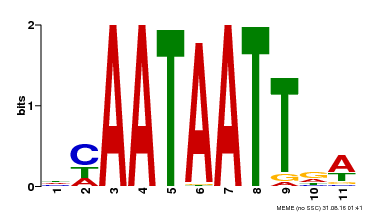
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011085824.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FQ391483 | 2e-68 | FQ391483.1 Vitis vinifera clone SS0AP1YK15. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011085824.1 | 0.0 | homeobox-leucine zipper protein HAT5 | ||||
| Swissprot | Q02283 | 2e-75 | HAT5_ARATH; Homeobox-leucine zipper protein HAT5 | ||||
| TrEMBL | A0A2G9G4U8 | 1e-126 | A0A2G9G4U8_9LAMI; Homeobox transcription factor | ||||
| STRING | Migut.H00128.1.p | 1e-123 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA7891 | 23 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 6e-76 | homeobox 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



