 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011086441.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 322aa MW: 35233.4 Da PI: 10.2221 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 41.2 | 3.7e-13 | 93 | 137 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+W++eE+ l++ + ++ G+g+W+ I+r k+Rt+ q+ s+ qky
XP_011086441.1 93 PWSEEEHRLFLLGLQKVGKGDWRGISRNYVKTRTPTQVASHAQKY 137
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.949 | 86 | 142 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.02E-16 | 88 | 143 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.8E-17 | 89 | 140 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 7.0E-10 | 90 | 140 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-10 | 92 | 137 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.6E-10 | 93 | 137 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.64E-9 | 93 | 138 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0080167 | Biological Process | response to karrikin | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003682 | Molecular Function | chromatin binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 322 aa Download sequence Send to blast |
MAAVQGGKCS DYSAMDDGRS SGTSSEVSGG KGFMLFGVRV MMEGSFRKSA SMNNLAQYDQ 60 QPQDSNNDVA SGYASDDVVH PSGRSRERKR GVPWSEEEHR LFLLGLQKVG KGDWRGISRN 120 YVKTRTPTQV ASHAQKYFLR RNNHNRRRRR SSLFDITTDA VVGSAIEDQM HQENSPQPPI 180 AQQQNLSKNL EKFPMSTYPV AITPLVVPIT SENHVENRTL GQVNQLNNSP RPIRPIPILP 240 VPPSSKMANL NLNKTTSLQP PSLSLNLSIS SQSPPPLPPS SQEPPPPPPS NQQSPPARHT 300 STFQAMPASF NESGGDSIIS VA |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00630 | PBM | Transfer from PK17526.1 | Download |
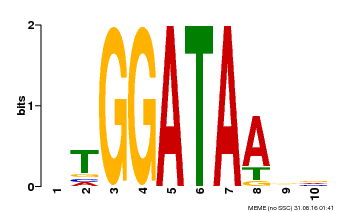
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011086441.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF008661 | 8e-84 | KF008661.1 Scutellaria baicalensis MYB12 (myb) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011086441.1 | 0.0 | transcription factor MYB1R1 | ||||
| TrEMBL | A0A2G9HKS2 | 1e-151 | A0A2G9HKS2_9LAMI; Uncharacterized protein | ||||
| STRING | Migut.L01371.1.p | 1e-128 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA11423 | 22 | 25 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G70000.2 | 8e-63 | MYB_related family protein | ||||



