 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011092666.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | NF-YB | ||||||||
| Protein Properties | Length: 141aa MW: 15879.8 Da PI: 5.7423 | ||||||||
| Description | NF-YB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NF-YB | 153.3 | 4.4e-48 | 3 | 98 | 2 | 97 |
NF-YB 2 reqdrflPianvsrimkkvlPanakiskdaketvqecvsefisfvtseasdkcqrekrktingddllwalatlGfedyveplkvylkkyrelegek 97
e+d++lPianv+rimk++lP akisk+aket+qec+sefi fvt+easd+c++e+rkt+ngdd++wal++lGf++y e++ yl+k re+e+++
XP_011092666.1 3 DEHDKLLPIANVGRIMKQILPPSAKISKEAKETMQECASEFICFVTGEASDRCHKENRKTVNGDDICWALSSLGFDHYSEAMLRYLQKLREFERQR 98
589******************************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.20.10 | 6.7E-48 | 2 | 117 | IPR009072 | Histone-fold |
| SuperFamily | SSF47113 | 9.47E-38 | 6 | 118 | IPR009072 | Histone-fold |
| Pfam | PF00808 | 5.1E-26 | 9 | 72 | IPR003958 | Transcription factor CBF/NF-Y/archaeal histone domain |
| PRINTS | PR00615 | 3.2E-16 | 36 | 54 | No hit | No description |
| PRINTS | PR00615 | 3.2E-16 | 55 | 73 | No hit | No description |
| PRINTS | PR00615 | 3.2E-16 | 74 | 92 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 141 aa Download sequence Send to blast |
MTDEHDKLLP IANVGRIMKQ ILPPSAKISK EAKETMQECA SEFICFVTGE ASDRCHKENR 60 KTVNGDDICW ALSSLGFDHY SEAMLRYLQK LREFERQRAN QSNYKPSSST EEKDEATSSA 120 ASAPSFDFEI LERGGRSSPK P |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4g91_B | 7e-38 | 4 | 93 | 3 | 92 | Transcription factor HapC (Eurofung) |
| 4g92_B | 7e-38 | 4 | 93 | 3 | 92 | Transcription factor HapC (Eurofung) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the NF-Y/HAP transcription factor complex. The NF-Y complex stimulates the transcription of various genes by recognizing and binding to a CCAAT motif in promoters. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00133 | DAP | Transfer from AT1G09030 | Download |
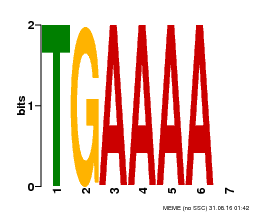
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011092666.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011092666.1 | 1e-102 | nuclear transcription factor Y subunit B-4 | ||||
| Swissprot | O04027 | 4e-52 | NFYB4_ARATH; Nuclear transcription factor Y subunit B-4 | ||||
| TrEMBL | A0A2G9IB77 | 1e-72 | A0A2G9IB77_9LAMI; CCAAT-binding factor, subunit A (HAP3) | ||||
| STRING | Migut.N01993.1.p | 7e-75 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA111 | 24 | 356 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09030.1 | 2e-54 | nuclear factor Y, subunit B4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



