 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011100397.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 847aa MW: 96659.1 Da PI: 6.8165 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 86.1 | 5.2e-27 | 90 | 192 | 2 | 91 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk.............tekerrtraetrtgCkaklkvkkekdgkwevtklel 83
fY+eYA++vGF++ +++s++sk+++e+++++f Cs++g+++e +k+ +e++ +ra +t+Cka+++vk+++dgkw ++++e+
XP_011100397.1 90 FYQEYARSVGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYEKSlnrprsrqgsnqdAENATGRRACAKTDCKASMHVKRRSDGKWIIHRFEK 184
9*****************************************999999***999988776677779***************************** PP
FAR1 84 eHnHelap 91
eHnHel p
XP_011100397.1 185 EHNHELLP 192
*****975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 5.7E-25 | 90 | 192 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 2.5E-27 | 290 | 382 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.458 | 570 | 606 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 3.0E-4 | 572 | 603 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 8.3E-8 | 581 | 608 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 847 aa Download sequence Send to blast |
MDIDLRLPSG GHDKEVEEEP NGIVNMLDGE EKPLNVDGVG GSMGDVEEKL QIEDAEDVNS 60 PIHDIDFKDV TILEPLPGME FGSHGDAYAF YQEYARSVGF NTAIQNSRRS KTSREFIDAK 120 FACSRYGTKR EYEKSLNRPR SRQGSNQDAE NATGRRACAK TDCKASMHVK RRSDGKWIIH 180 RFEKEHNHEL LPAQAVSEQT RRMYAAMARQ FAEYKSAVGL KHDSRSHFEK GRNMAMDAGE 240 ANMLLDFFVQ MQSLNSNFFY AVDVGEDQRL KNLLWIDAKS RHDYPNFSDV VSFDTSYVRN 300 KYKMPLALFV GVNQHYQFML LGCALVSDES EATFSWVMQT WLKAMGGQAP KIIITDQDKV 360 MKSVTADAFP STLHFFCLWN IMGKVSETLN HVIKQNENFM SKFEKCVYRS WTDDEFDKRW 420 HKLVNRFGLK ENELMQSLYE DRKKWVPNFM KDGFFAGMST GQRSESVNSF FDKYVHKKTT 480 MQEFIKQYEA ILQDRYEEEA KASSDTWNKQ PSLKSPSPFE KHVAGLYTHA VFRKFQVEVL 540 GAVACIPKRE EQVDATITFK VQDFERNQEF IVTLNELKSE VSCICRLFEF KGFLCRHAMI 600 VLQICGISNI PSQYILKRWT KDAKSRYPMG EGSEQVQSRL QRYNDLCQRA IKLGEEGSFS 660 QESYNLTLRA LDDAFETCLN ANNSCKNLLE AGPSSSPALL CIEEDIQSGN LSKTNKKKSS 720 TKKRKVNMEP DVITVGTPES LQQMEKLNSR PVNLDGFFGA QQGVQGMVQL NLMAPTRDNY 780 YGNQQTIQGL GQLNSIAPTH DGYYGTQPAI HGLGQMDFFR TPSFGYGIRE DPNVRSAQLH 840 DDAPRHA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
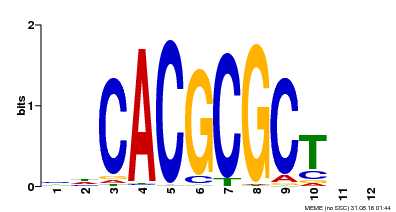
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011100397.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011100397.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X2 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A2G9GLI3 | 0.0 | A0A2G9GLI3_9LAMI; Uncharacterized protein | ||||
| STRING | Migut.M00094.1.p | 0.0 | (Erythranthe guttata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



