 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011100790.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 927aa MW: 104925 Da PI: 7.2512 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 165.9 | 6.6e-52 | 30 | 147 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
++e k+rwl+++ei+aiL n++ +++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+nptf
XP_011100790.1 30 MEEaKARWLRPNEIHAILCNHKYFTVYVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNEERIHVYYAHGEDNPTFV 124
45669****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcywlL+++le+ivlvhy+e++
XP_011100790.1 125 RRCYWLLDKSLEHIVLVHYRETQ 147
********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 77.671 | 26 | 152 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.5E-77 | 29 | 147 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 7.0E-46 | 32 | 145 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.22E-9 | 380 | 465 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.30E-13 | 564 | 674 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 5.1E-15 | 565 | 677 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.42E-16 | 565 | 677 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.345 | 573 | 647 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 5.1E-6 | 615 | 644 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.434 | 615 | 647 | IPR002110 | Ankyrin repeat |
| Pfam | PF00023 | 9.2E-5 | 616 | 646 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.92E-6 | 723 | 829 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.163 | 763 | 789 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 14 | 778 | 800 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.089 | 782 | 808 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0012 | 801 | 823 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.091 | 802 | 826 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.001 | 803 | 823 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 35 | 881 | 903 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.114 | 883 | 911 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 927 aa Download sequence Send to blast |
MENTGVRGRF VGSEIHGFRT LEDLDVGNMM EEAKARWLRP NEIHAILCNH KYFTVYVKPV 60 NLPKSGTIVL FDRKMLRNFR KDGHNWKKKK DGKTVKEAHE HLKVGNEERI HVYYAHGEDN 120 PTFVRRCYWL LDKSLEHIVL VHYRETQEGS PATPVNSNSN SAGSDLSATW PMSEESDSAV 180 DRVYYGSTGS YLECHDSVTV KHHEQRLYEI NTLEWDELLV PDDPHRLITR QQGTTAGFEL 240 QNQYQMNSYR INDDAPSNNK VSPECSTNSF SEPVAGRSSI NYTSPNNMSY QTVEQDTIVN 300 SETMVSGLMP SGGAGSLYNL GKDGLQSQDS FGRWVTHIIA ESPESVDDHT LESSNLAGHQ 360 SSTYPLMDSH DSSPLGPIFT ITDVSPAWAL STEETKILVV GFFNEGQLPY SESKLYLACG 420 DSLLPVDVVQ AGVFRCLIPP QAPKLGNLYI TFDGHKPISQ VLTFEIRAPV QPGTVSFENK 480 TDWEEFQLQM RLAHLLFSSS KGLSIYSTKL SPTALKEAKA FAQKTSHISD GWLHMAKVIE 540 DTKMSFPQAK DKLFELTLQN RLQEWLLEKV VAGCKISERD EQGLGVIHLC SILGYTWAVY 600 PYSWSGLSLD YRDKFGWTAL HWAAYYGREK MVATLLSAGA KPNLVTDPTS QNPGGCSAHD 660 LASKNGYDGL AAYLAEKALV AQFDDMTLAG NVSGSLQTTT NETVNPGNFS EDELYLKDTL 720 AAYRTAADAA ARIQTAFREH SLKIRTKVVE SSNPELEARN IVAAMKIQHA FRNYETRKKI 780 VAAARIQHRF RTWKIRKEFL NMRRQAIKIQ AMFRGFQVRR QYRKIVWSVG VLEKAILRWR 840 LKRKGFRGLQ VQPAETPREP NEESDVEEDF FQASRKQAEE RVEQSVVRVQ AMFRSKQAQE 900 AYRRMKLEHN KAKLEYEGLL HPDLQMG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
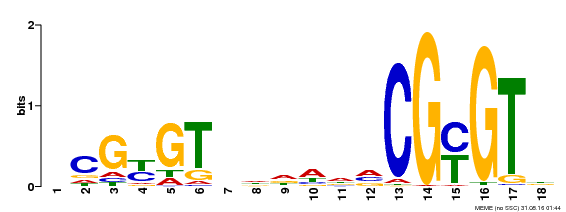
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011100790.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011100790.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A022RYH3 | 0.0 | A0A022RYH3_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.G00093.1.p | 0.0 | (Erythranthe guttata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



