 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011100793.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 875aa MW: 98969.6 Da PI: 7.2089 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 137.6 | 3.9e-43 | 10 | 93 | 35 | 118 |
CG-1 35 ksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
+sg+++L++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+nptf rrcywlL+++le+ivlvhy+e++
XP_011100793.1 10 TSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNEERIHVYYAHGEDNPTFVRRCYWLLDKSLEHIVLVHYRETQ 93
79*******************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 61.923 | 1 | 98 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 9.5E-48 | 1 | 93 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.3E-36 | 10 | 91 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 2.99E-9 | 328 | 413 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.19E-13 | 512 | 622 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 4.7E-15 | 513 | 625 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.11E-16 | 513 | 626 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.345 | 521 | 595 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.434 | 563 | 595 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 5.1E-6 | 563 | 592 | IPR002110 | Ankyrin repeat |
| Pfam | PF00023 | 8.6E-5 | 564 | 594 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.04E-6 | 670 | 777 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.163 | 711 | 737 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 14 | 726 | 748 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.089 | 730 | 756 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0012 | 749 | 771 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.091 | 750 | 774 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 9.4E-4 | 751 | 771 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 35 | 829 | 851 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.114 | 831 | 859 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 875 aa Download sequence Send to blast |
MRFMPYSVTT SGTIVLFDRK MLRNFRKDGH NWKKKKDGKT VKEAHEHLKV GNEERIHVYY 60 AHGEDNPTFV RRCYWLLDKS LEHIVLVHYR ETQELQGSPA TPVNSNSNSA GSDLSATWPM 120 SEESDSAVDR VYYGSTGSYL ECHDSVTVKH HEQRLYEINT LEWDELLVPD DPHRLITRQQ 180 GTTAGFELQN QYQMNSYRIN DDAPSNNKVS PECSTNSFSE PVAGRSSINY TSPNNMSYQT 240 VEQDTIVNSE TMVSGLMPSG GAGSLYNLGK DGLQSQDSFG RWVTHIIAES PESVDDHTLE 300 SSNLAGHQSS TYPLMDSHDS SPLGPIFTIT DVSPAWALST EETKILVVGF FNEGQLPYSE 360 SKLYLACGDS LLPVDVVQAG VFRCLIPPQA PKLGNLYITF DGHKPISQVL TFEIRAPVQP 420 GTVSFENKTD WEEFQLQMRL AHLLFSSSKG LSIYSTKLSP TALKEAKAFA QKTSHISDGW 480 LHMAKVIEDT KMSFPQAKDK LFELTLQNRL QEWLLEKVVA GCKISERDEQ GLGVIHLCSI 540 LGYTWAVYPY SWSGLSLDYR DKFGWTALHW AAYYGREKMV ATLLSAGAKP NLVTDPTSQN 600 PGGCSAHDLA SKNGYDGLAA YLAEKALVAQ FDDMTLAGNV SGSLQTTTNE TVNPGNFSED 660 ELYLKDTLAA YRTAADAAAR IQTAFREHSL KIRTKVVESS NPELEARNIV AAMKIQHAFR 720 NYETRKKIVA AARIQHRFRT WKIRKEFLNM RRQAIKIQAM FRGFQVRRQY RKIVWSVGVL 780 EKAILRWRLK RKGFRGLQVQ PAETPREPNE ESDVEEDFFQ ASRKQAEERV EQSVVRVQAM 840 FRSKQAQEAY RRMKLEHNKA KLEYEGLLHP DLQMG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
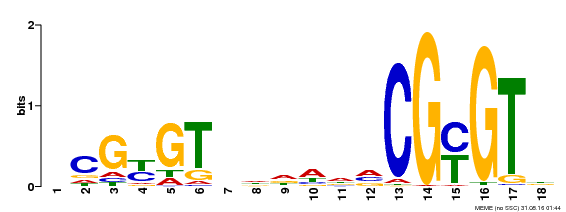
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_011100793.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011100793.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X4 | ||||
| Refseq | XP_011100794.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X4 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A022RYH3 | 0.0 | A0A022RYH3_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.G00093.1.p | 0.0 | (Erythranthe guttata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



