 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011399305.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Chlorellales; Chlorellaceae; Auxenochlorella
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 198aa MW: 20332.8 Da PI: 6.0028 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 98.3 | 5.4e-31 | 93 | 147 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k rlrWtp LH++F +av+qLGG+ekAtPk il+lm v gLt++h+kSHLQkYRl
XP_011399305.1 93 KSRLRWTPSLHASFARAVSQLGGPEKATPKGILKLMRVSGLTIYHIKSHLQKYRL 147
68****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.627 | 90 | 150 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-29 | 91 | 149 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 8.42E-17 | 92 | 150 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-23 | 93 | 147 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.4E-8 | 95 | 146 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 198 aa Download sequence Send to blast |
MKGPDRDLLL NSLLEPSWAQ ASTSATGTGW TGGLGPGPGS SAPGVAAALH PYGVYLHPLA 60 LPIGGSPASS TAPGMSDGPD NFKFEDSGPS NSKSRLRWTP SLHASFARAV SQLGGPEKAT 120 PKGILKLMRV SGLTIYHIKS HLQKYRLSIK LPGEGGDGED EAEGEQQRLG LAGDSEDDCG 180 RPLEPAALDP ALALASAA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 6e-24 | 93 | 146 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-24 | 93 | 146 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-24 | 93 | 146 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-24 | 93 | 146 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
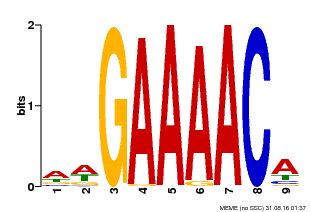
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011399305.1 | 1e-139 | Myb family transcription factor APL | ||||
| TrEMBL | A0A087SL05 | 1e-137 | A0A087SL05_AUXPR; Myb family transcription factor APL | ||||
| STRING | A0A087SL05 | 1e-138 | (Auxenochlorella protothecoides) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP111 | 16 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 6e-26 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 23616293 |



