 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_011400581.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Chlorellales; Chlorellaceae; Auxenochlorella
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 282aa MW: 29025 Da PI: 7.0418 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 33 | 1.1e-10 | 139 | 184 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
h ++E rRR+riN+++e Lre++P++ ++ + a+ Le + +Y++ +q
XP_011400581.1 139 HQQAEARRRSRINDRLEALREVVPHS-----ERANTATFLEDVLKYVRAMQ 184
899**********************9.....8***************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 12.627 | 134 | 183 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-12 | 137 | 194 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.75E-11 | 137 | 199 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.05E-8 | 137 | 185 | No hit | No description |
| Pfam | PF00010 | 6.7E-8 | 138 | 184 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.2E-4 | 140 | 189 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 282 aa Download sequence Send to blast |
MDYPPRNPGE AGDPGQAQPD SGQGGMNPAT LQALLAASAR SSAFGPVGGQ LPHPPPFPTE 60 PGPGQQLGPS SMHLFNQLLG QPGLAHLAGY PDSGPPSGPG PAPGNAPPSP TSGGDGGGGK 120 KQRTRAGPSP SSSSYASRHQ QAEARRRSRI NDRLEALREV VPHSERANTA TFLEDVLKYV 180 RAMQRRMAEL EAQQPHAWPP GMQAAAAQQQ QRGMGHMEQD PGAMFGAQAG QPAAQLAQAQ 240 AAHAAFAAHA QAQQAAASGL AGSNEEEERA KQAQEVGAQA PL |
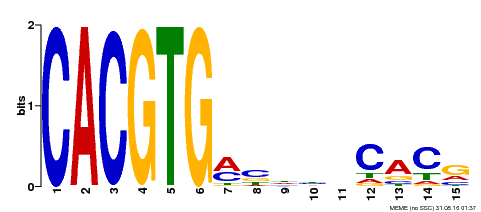
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011400581.1 | 0.0 | Transcription factor BIM2 | ||||
| TrEMBL | A0A087SPE4 | 0.0 | A0A087SPE4_AUXPR; Transcription factor BIM2 | ||||
| STRING | A0A087SPE4 | 0.0 | (Auxenochlorella protothecoides) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP5665 | 8 | 8 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69010.1 | 4e-11 | BES1-interacting Myc-like protein 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 23612248 |



