 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_012574211.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 808aa MW: 89088.2 Da PI: 6.1868 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.2 | 4.5e-21 | 118 | 173 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
XP_012574211.1 118 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 173
688999***********************************************999 PP
| |||||||
| 2 | START | 210.2 | 7.6e-66 | 320 | 544 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetl 82
ela++a++elvk+a+ +ep+W +s e++n +e++++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++ + +t+
XP_012574211.1 320 ELALAAMDELVKMAQTSEPLWIRSIeggrEILNHEEYMRTFTPCIGlrpngFVSEASRETGMVIINSLALVETLMDSN-RWIEMFPciiaRTSTT 413
5899**************************************9999********************************.**************** PP
EEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-E CS
START 83 evissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehv 170
evis+g galqlm+ael +lsplvp R++ f+R+++q+ +g+w++vdvS+ds ++++ +s+v +++lpSg+++++++ng+skvtwveh+
XP_012574211.1 414 EVISNGingtrnGALQLMQAELHVLSPLVPvREVNFLRFCKQHAEGVWAVVDVSIDSIRENSGAPSFVNCRKLPSGCVVQDMPNGYSKVTWVEHA 508
**************************************************************9******************************** PP
E--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 171 dlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+++++++h+l+r+l++sg+ +ga +wv tlqrqce+
XP_012574211.1 509 EYEENQVHQLYRPLLSSGMGFGATRWVVTLQRQCEC 544
**********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.54E-20 | 103 | 175 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.8E-22 | 107 | 175 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 115 | 175 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.9E-18 | 116 | 179 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.93E-18 | 117 | 175 | No hit | No description |
| Pfam | PF00046 | 1.4E-18 | 118 | 173 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 150 | 173 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.721 | 311 | 547 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.65E-34 | 313 | 544 | No hit | No description |
| CDD | cd08875 | 5.91E-127 | 315 | 543 | No hit | No description |
| Pfam | PF01852 | 2.1E-57 | 320 | 544 | IPR002913 | START domain |
| SMART | SM00234 | 1.8E-48 | 320 | 544 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.2E-23 | 572 | 801 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 808 aa Download sequence Send to blast |
MSFGGFVENN SGGGSVRNIA AEISYNNNQR MSFGSISHPR LVTTPTLAKS MFNSPGLSLA 60 LQQTNIDGQE DVNRSMHENF EQNGLRRSRE EEQSRSGSDN LDGVSGDEQD ADDKPPRKKR 120 YHRHTPQQIQ ELEALFKECP HPDEKQRLEL SKRLCLETRQ VKFWFQNRRT QMKTQLERHE 180 NSLLRQENDK LRAENMSIRD AMRNPICSNC GGPAMIGEIS LEEQHLRIEN ARLKDELDRV 240 CALAGKFLGR PISTLPNSSL ELGVGGNNGF NGMNNVSSTL PDFGVGMSNN PLAIVSPSTR 300 QTTPLVTGFD RSVERSMFLE LALAAMDELV KMAQTSEPLW IRSIEGGREI LNHEEYMRTF 360 TPCIGLRPNG FVSEASRETG MVIINSLALV ETLMDSNRWI EMFPCIIART STTEVISNGI 420 NGTRNGALQL MQAELHVLSP LVPVREVNFL RFCKQHAEGV WAVVDVSIDS IRENSGAPSF 480 VNCRKLPSGC VVQDMPNGYS KVTWVEHAEY EENQVHQLYR PLLSSGMGFG ATRWVVTLQR 540 QCECLAILMS SAAPSRDHSA ITAGGRRSML KLAQRMTNNF CAGVCASTVH KWNKLSPGNV 600 DEDVRVMTRK XXXDPGEPPG IVLSAATSVW LPVSPQRLFD FLRDERLRSE WDILSNGGPM 660 QEMAHIAKGQ DHGNCVSLLR ASAMNSNQSS MLILQETCID EAGSLVVYAP VDIPAMHVVM 720 NGGDSAYVAL LPSGFAVVPD GPGSRGPENE TTTNGGETRV SGSLLTVAFQ ILVNSLPTAK 780 LTVESVETVN NLISCTVQKI KAALQCES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
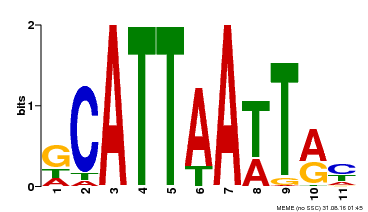
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_012574211.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012574211.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2-like isoform X1 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A1S3EGX5 | 0.0 | A0A1S3EGX5_CICAR; homeobox-leucine zipper protein ANTHOCYANINLESS 2-like isoform X1 | ||||
| STRING | XP_004510857.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1192 | 34 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



