 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013583747.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 289aa MW: 31620.2 Da PI: 9.3792 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.5 | 5.6e-12 | 5 | 56 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+ +WT++E++ l+ +v+++G+g Wk I r R+ ++k++w+++
XP_013583747.1 5 KSKWTEDEEDALLAGVRKHGPGKWKNILRDPEfagvlSLRSNIDLKDKWRNL 56
679**********************************88***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.218 | 1 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.02E-15 | 2 | 61 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.2E-14 | 3 | 57 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.0E-8 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-8 | 5 | 56 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 3.30E-18 | 7 | 56 | No hit | No description |
| SMART | SM00526 | 1.4E-14 | 124 | 188 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 1.8E-12 | 125 | 197 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 1.4E-8 | 126 | 187 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PROSITE profile | PS51504 | 19.756 | 126 | 193 | IPR005818 | Linker histone H1/H5, domain H15 |
| Pfam | PF00538 | 4.8E-9 | 128 | 186 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 289 aa Download sequence Send to blast |
MGKEKSKWTE DEEDALLAGV RKHGPGKWKN ILRDPEFAGV LSLRSNIDLK DKWRNLSVPA 60 DIQGSKDKVR TPKIKAAAFQ LAAVAAAATT PSPSSVSSPV APPLPRSGSS DLSIGDSCNM 120 LIDAKNAPRY DGMIFEALSA LKDANGSDVT TIFNFIEQKM YEVPEKRVLG SRLRRLAAQG 180 KLEKTQNLYR MNNNSFLTMR TPVPAARPKE ANAKPRNSQG LTVSQEKIAL ASGTAAFKLY 240 ELDGKLEVLG GAVDDRARMT ELAERAEIML LLAEELHERC CRGEIVELN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
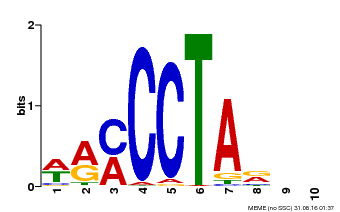
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013583747.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013583747.1 | 0.0 | PREDICTED: telomere repeat-binding factor 4 isoform X2 | ||||
| Refseq | XP_013744609.1 | 0.0 | telomere repeat-binding factor 4 isoform X2 | ||||
| Swissprot | F4I7L1 | 1e-108 | TRB4_ARATH; Telomere repeat-binding factor 4 | ||||
| TrEMBL | A0A078IBP9 | 0.0 | A0A078IBP9_BRANA; BnaC05g13640D protein | ||||
| TrEMBL | A0A0D3CB23 | 0.0 | A0A0D3CB23_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6ESL4 | 0.0 | A0A3P6ESL4_BRAOL; Uncharacterized protein | ||||
| STRING | Bo5g023860.1 | 0.0 | (Brassica oleracea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G17520.1 | 1e-107 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



