 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013588453.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 298aa MW: 32738.4 Da PI: 8.2019 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 101.7 | 5.1e-32 | 64 | 119 | 2 | 58 |
ZF-HD_dimer 2 ekvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRreve 58
++vrY+eClkNhAa++Gg + DGCgEfmps geegt++al+CaAC+CHRnFHR+ev+
XP_013588453.1 64 TAVRYRECLKNHAANVGGSVHDGCGEFMPS-GEEGTIEALRCAACDCHRNFHRKEVD 119
579**************************9.999********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 2.0E-27 | 60 | 119 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 2.9E-29 | 66 | 118 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 5.1E-30 | 67 | 118 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 26.03 | 68 | 117 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| SuperFamily | SSF46689 | 5.87E-20 | 221 | 292 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-31 | 223 | 292 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 8.746 | 229 | 293 | IPR001356 | Homeobox domain |
| TIGRFAMs | TIGR01565 | 8.6E-30 | 231 | 288 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 298 aa Download sequence Send to blast |
MDMRSHEMIE RRRDDNDDNG VGSIANEDNV NGNHNNTRVS SQTLDHHHQS KSPSSFAISA 60 AAKTAVRYRE CLKNHAANVG GSVHDGCGEF MPSGEEGTIE ALRCAACDCH RNFHRKEVDG 120 VGSSDVIAHH HRHHHHQYGG GGGGRRPPPP NMMVNPLMLP PPPNYAPMHH HKYGMSPPGG 180 AGMVTPMSVA YGGSGGGGAE SSSEDLNMYG QSSGEHGGAA AGQMAFSMSS KKRFRTKFTT 240 EQKERMMEFA EKLGWRMNKQ DEEELKRFCD EIGVKRQVFK VWMHNNKNNA RKPPPSTV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh7_A | 1e-22 | 230 | 290 | 16 | 76 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Mostly expressed in flowers and inflorescence. {ECO:0000269|PubMed:16428600}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
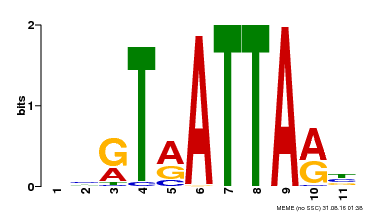
| UniProt | Putative transcription factor. Binds DNA at 5'-ATTA-3' consensus promoter regions. Regulates floral architecture and leaf development. Regulators in the abscisic acid (ABA) signal pathway that confers sensitivity to ABA in an ARF2-dependent manner. {ECO:0000269|PubMed:16428600, ECO:0000269|PubMed:21059647, ECO:0000269|PubMed:21779177}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00238 | DAP | Transfer from AT1G75240 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013588453.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by abscisic acid (ABA) and ARF2. {ECO:0000269|PubMed:21779177}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC189493 | 0.0 | AC189493.2 Brassica rapa subsp. pekinensis cultivar Inbred line 'Chiifu' clone KBrB084P16, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013588453.1 | 0.0 | PREDICTED: zinc-finger homeodomain protein 5 | ||||
| Swissprot | Q9FRL5 | 1e-119 | ZHD5_ARATH; Zinc-finger homeodomain protein 5 | ||||
| TrEMBL | A0A0D3D0U3 | 0.0 | A0A0D3D0U3_BRAOL; Uncharacterized protein | ||||
| STRING | Bo6g119380.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7279 | 25 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G75240.1 | 1e-105 | homeobox protein 33 | ||||



