 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013589953.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 288aa MW: 32493.3 Da PI: 5.6061 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57 | 3.4e-18 | 79 | 132 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rq+ +WFqNrRa++k
XP_013589953.1 79 KKRRLNMEQVKTLEKTFELGNKLEPEMKMQLARALGLQPRQIAIWFQNRRARWK 132
456899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.7 | 1.2e-41 | 78 | 169 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
ekkrrl+ eqvk+LE++Fe +kLepe K++lar+Lglqprq+a+WFqnrRAR+ktkqlEkdy++Lk+++dalk++ne L++++++L++e++
XP_013589953.1 78 EKKRRLNMEQVKTLEKTFELGNKLEPEMKMQLARALGLQPRQIAIWFQNRRARWKTKQLEKDYDTLKQQFDALKADNELLQTHNQKLQAEIM 169
69**************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.37E-19 | 66 | 136 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.73 | 74 | 134 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.2E-18 | 77 | 138 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.96E-17 | 79 | 135 | No hit | No description |
| Pfam | PF00046 | 1.4E-15 | 79 | 132 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-19 | 86 | 133 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.3E-5 | 105 | 114 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 109 | 132 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.3E-5 | 114 | 130 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.8E-16 | 134 | 176 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 288 aa Download sequence Send to blast |
MSCNNGMSFF PSNFMIQTSY EDDHPHQAPS LAPLLPSCSI PQDLHGFASF LGKRSPIEVG 60 NNMNGEEDYS DDGSHMGEKK RRLNMEQVKT LEKTFELGNK LEPEMKMQLA RALGLQPRQI 120 AIWFQNRRAR WKTKQLEKDY DTLKQQFDAL KADNELLQTH NQKLQAEIMG LKNKEQIESI 180 NLNKETEGSC SNRSDNSSDN LRLDISTALP SVDSTITGGH PPAPQTVGRH FFPPSPAAAT 240 TTTTTMQFFQ NSSSGQSMVK EENSISNMLC AMDDHSGFWP WLDQQQYN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 126 | 134 | RRARWKTKQ |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Predominantly expressed in leaves and flowers. {ECO:0000269|PubMed:16055682}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may act in the sucrose-signaling pathway. {ECO:0000269|PubMed:11292072}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
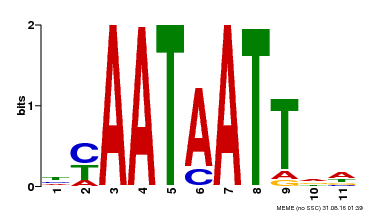
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013589953.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353311 | 0.0 | AK353311.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-34-O11. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009105645.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-13 | ||||
| Refseq | XP_013589953.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-13 | ||||
| Refseq | XP_013698957.1 | 0.0 | homeobox-leucine zipper protein ATHB-13 | ||||
| Refseq | XP_013728836.1 | 0.0 | homeobox-leucine zipper protein ATHB-13 | ||||
| Swissprot | Q8LC03 | 0.0 | ATB13_ARATH; Homeobox-leucine zipper protein ATHB-13 | ||||
| TrEMBL | A0A0D3CZD9 | 0.0 | A0A0D3CZD9_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6FZV1 | 0.0 | A0A3P6FZV1_BRAOL; Uncharacterized protein | ||||
| TrEMBL | M4DID9 | 0.0 | M4DID9_BRARP; Uncharacterized protein | ||||
| STRING | Bra016266.1-P | 0.0 | (Brassica rapa) | ||||
| STRING | Bo6g110380.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2633 | 27 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



